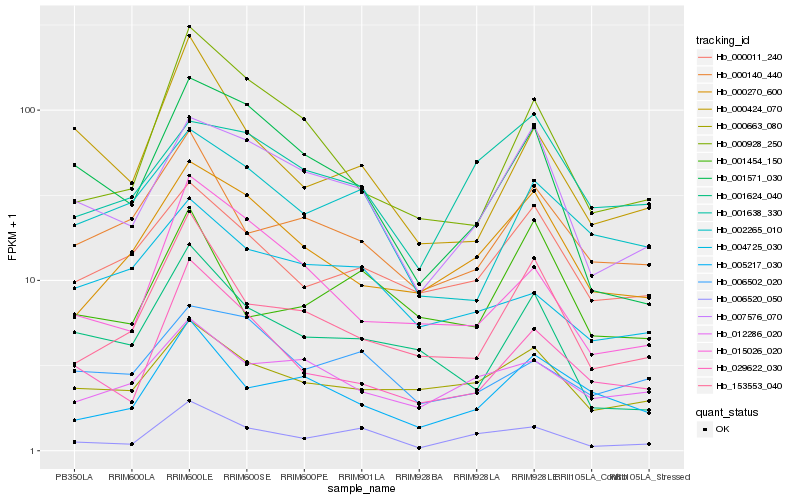
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006520_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007576_070 |
0.1718723403 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X3 [Jatropha curcas] |
| 3 |
Hb_029622_030 |
0.1802966699 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_153553_040 |
0.1846813736 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 5 |
Hb_000663_080 |
0.1866323555 |
transcription factor |
TF Family: HB |
Homeobox protein HAT3.1, putative [Ricinus communis] |
| 6 |
Hb_001571_030 |
0.190407288 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000011_240 |
0.1917665549 |
- |
- |
PREDICTED: uncharacterized protein LOC105631316 [Jatropha curcas] |
| 8 |
Hb_000270_600 |
0.1927843292 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006502_020 |
0.1934722803 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 10 |
Hb_012286_020 |
0.1965337523 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 11 |
Hb_015026_020 |
0.1966551698 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 12 |
Hb_000424_070 |
0.1977466139 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 13 |
Hb_000140_440 |
0.1984455896 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 14 |
Hb_005217_030 |
0.1998309992 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 15 |
Hb_004725_030 |
0.2005353275 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 16 |
Hb_001454_150 |
0.2024832649 |
- |
- |
isoleucyl tRNA synthetase, putative [Ricinus communis] |
| 17 |
Hb_001638_330 |
0.2033592041 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001624_040 |
0.2044028341 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000928_250 |
0.2059282413 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_002265_010 |
0.2061119832 |
- |
- |
ef-hand calcium binding protein, putative [Ricinus communis] |