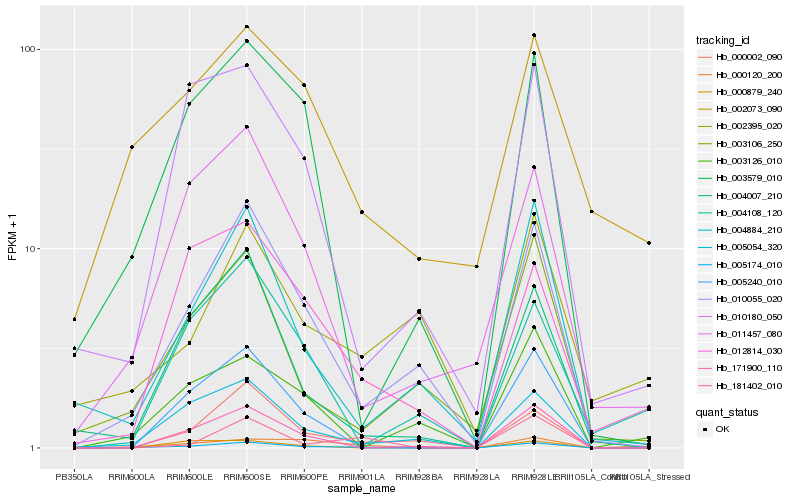
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005174_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_010055_020 |
0.2418185923 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 3 |
Hb_012814_030 |
0.2472133524 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 4 |
Hb_002073_090 |
0.2509875065 |
- |
- |
PREDICTED: copper transporter 6-like [Jatropha curcas] |
| 5 |
Hb_011457_080 |
0.2514817994 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004108_120 |
0.2540604402 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |
| 7 |
Hb_000120_200 |
0.2550559411 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 8 |
Hb_005054_320 |
0.2598501401 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 9 |
Hb_005240_010 |
0.2643343349 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 10 |
Hb_004007_210 |
0.2644929447 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 11 |
Hb_003126_010 |
0.2660373056 |
- |
- |
PREDICTED: oligopeptide transporter 7-like isoform X1 [Malus domestica] |
| 12 |
Hb_004884_210 |
0.2671790011 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 13 |
Hb_003579_010 |
0.2694970933 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 14 |
Hb_000002_090 |
0.2700195285 |
- |
- |
PREDICTED: RING-H2 finger protein ATL8-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_003106_250 |
0.2712717133 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 16 |
Hb_000879_240 |
0.2712952311 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631008 [Jatropha curcas] |
| 17 |
Hb_181402_010 |
0.2720050675 |
- |
- |
PREDICTED: uncharacterized protein LOC104905886 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_171900_110 |
0.2723599876 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 19 |
Hb_010180_050 |
0.272747293 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 20 |
Hb_002395_020 |
0.2728908285 |
- |
- |
Protein CREG1 precursor, putative [Ricinus communis] |