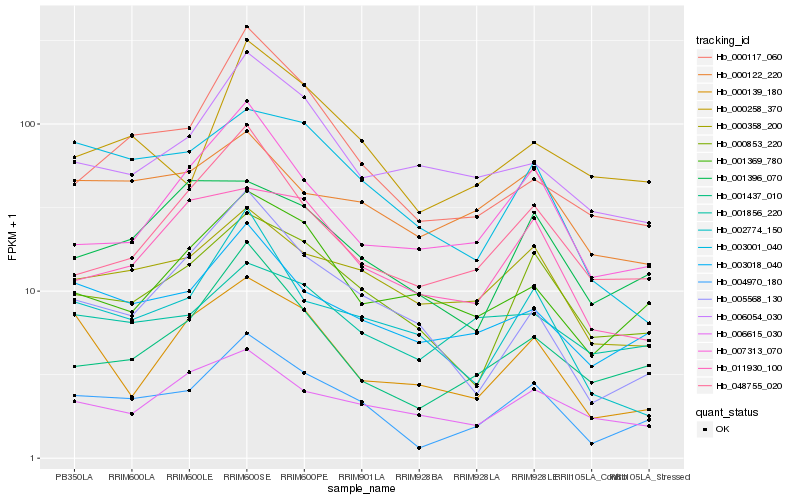
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004970_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_003018_040 |
0.1382514353 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000853_220 |
0.1389791685 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 4 |
Hb_001437_010 |
0.1427708525 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 5 |
Hb_000358_200 |
0.1516723275 |
- |
- |
PREDICTED: uncharacterized protein LOC105637771 [Jatropha curcas] |
| 6 |
Hb_000258_370 |
0.1577331224 |
- |
- |
hypothetical protein [Ricinus communis] |
| 7 |
Hb_048755_020 |
0.1622182881 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 8 |
Hb_007313_070 |
0.1647725978 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 9 |
Hb_003001_040 |
0.1675725995 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 10 |
Hb_001369_780 |
0.1713866112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002774_150 |
0.1725162872 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 12 |
Hb_000122_220 |
0.1737208629 |
- |
- |
Prenylated Rab acceptor protein, putative [Ricinus communis] |
| 13 |
Hb_000117_060 |
0.1740153828 |
- |
- |
PREDICTED: uncharacterized protein LOC103949199 [Pyrus x bretschneideri] |
| 14 |
Hb_001856_220 |
0.1742016982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001396_070 |
0.1760922505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011930_100 |
0.1762796364 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 17 |
Hb_005568_130 |
0.1765814601 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000139_180 |
0.1766448074 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 19 |
Hb_006615_030 |
0.1771512148 |
- |
- |
prolyl oligopeptidase family protein [Populus trichocarpa] |
| 20 |
Hb_006054_030 |
0.1774071364 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |