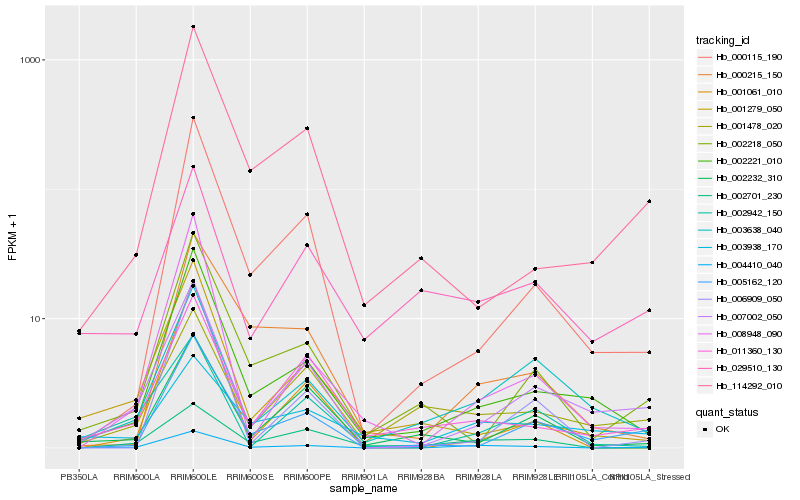
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004410_040 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_007002_050 |
0.2109859302 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 3 |
Hb_002221_010 |
0.2151732241 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3-like [Jatropha curcas] |
| 4 |
Hb_002942_150 |
0.2309604801 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 5 |
Hb_011360_130 |
0.2318878268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008948_090 |
0.2387674005 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000115_190 |
0.2388816998 |
- |
- |
PREDICTED: uncharacterized protein LOC105642039 [Jatropha curcas] |
| 8 |
Hb_003638_040 |
0.2393184884 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 9 |
Hb_002701_230 |
0.2453357723 |
- |
- |
PREDICTED: uncharacterized protein LOC105646833 [Jatropha curcas] |
| 10 |
Hb_002218_050 |
0.2461522911 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 11 |
Hb_001279_050 |
0.2466123437 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 12 |
Hb_005162_120 |
0.2501289149 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 13 |
Hb_000215_150 |
0.2510170729 |
- |
- |
PREDICTED: uncharacterized protein LOC100249186 [Vitis vinifera] |
| 14 |
Hb_003938_170 |
0.252548169 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 15 |
Hb_001061_010 |
0.2556432987 |
- |
- |
PREDICTED: uncharacterized protein LOC100243595 [Vitis vinifera] |
| 16 |
Hb_001478_020 |
0.2583713626 |
desease resistance |
Gene Name: AAA |
thyroid hormone receptor interactor, putative [Ricinus communis] |
| 17 |
Hb_002232_310 |
0.2595729873 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 18 |
Hb_029510_130 |
0.2609665113 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 19 |
Hb_006909_050 |
0.2629531682 |
transcription factor |
TF Family: OFP |
hypothetical protein POPTR_0008s01840g [Populus trichocarpa] |
| 20 |
Hb_114292_010 |
0.2639622662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |