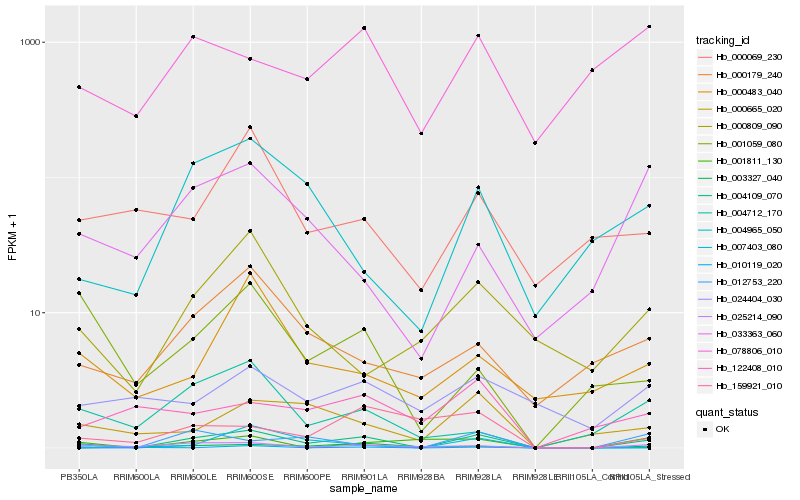
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004109_070 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa010934mg [Prunus persica] |
| 2 |
Hb_025214_090 |
0.2979452535 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 3 |
Hb_010119_020 |
0.3208248232 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_004965_050 |
0.3250980961 |
- |
- |
PREDICTED: uncharacterized protein LOC105645055 [Jatropha curcas] |
| 5 |
Hb_004712_170 |
0.3259216758 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 6 |
Hb_000665_020 |
0.3324821302 |
- |
- |
- |
| 7 |
Hb_000179_240 |
0.3398862583 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003327_040 |
0.344656676 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 9 |
Hb_000809_090 |
0.347823358 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 10 |
Hb_078806_010 |
0.3478967578 |
- |
- |
hypothetical protein CISIN_1g031839mg [Citrus sinensis] |
| 11 |
Hb_033363_060 |
0.3569721515 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 12 |
Hb_007403_080 |
0.3577978367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001811_130 |
0.3588845517 |
- |
- |
- |
| 14 |
Hb_000069_230 |
0.3614450282 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 15 |
Hb_001059_080 |
0.3623228851 |
- |
- |
PREDICTED: F-box protein At1g61340-like [Musa acuminata subsp. malaccensis] |
| 16 |
Hb_000483_040 |
0.3625099503 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 17 |
Hb_122408_010 |
0.365436929 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_024404_030 |
0.3656453832 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14580, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_012753_220 |
0.3670586499 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 20 |
Hb_159921_010 |
0.3673012121 |
- |
- |
- |