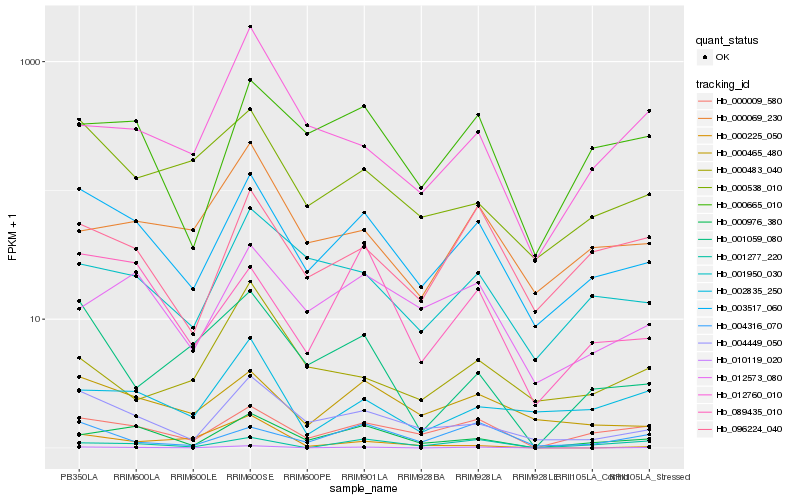
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010119_020 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_003517_060 |
0.2382436061 |
- |
- |
hypothetical protein CISIN_1g044999mg, partial [Citrus sinensis] |
| 3 |
Hb_000976_380 |
0.2602358035 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_004449_050 |
0.2604745099 |
- |
- |
PREDICTED: tyrosine-sulfated glycopeptide receptor 1-like [Jatropha curcas] |
| 5 |
Hb_001059_080 |
0.2704255949 |
- |
- |
PREDICTED: F-box protein At1g61340-like [Musa acuminata subsp. malaccensis] |
| 6 |
Hb_000009_580 |
0.2732120145 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 7 |
Hb_012760_010 |
0.2808240628 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000069_230 |
0.281002426 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 9 |
Hb_004316_070 |
0.2945672427 |
- |
- |
E3 ubiquitin-protein ligase UPL5 -like protein [Gossypium arboreum] |
| 10 |
Hb_002835_250 |
0.2962047306 |
- |
- |
octicosapeptide/Phox/Bem1p domain-containing family protein [Populus trichocarpa] |
| 11 |
Hb_000665_010 |
0.2971992819 |
- |
- |
hypothetical protein 20 [Hevea brasiliensis] |
| 12 |
Hb_001950_030 |
0.2978782432 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 13 |
Hb_096224_040 |
0.2985184557 |
- |
- |
PREDICTED: probable nucleoredoxin 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000225_050 |
0.3004946728 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000483_040 |
0.3011943594 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 16 |
Hb_089435_010 |
0.3035410221 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Vitis vinifera] |
| 17 |
Hb_001277_220 |
0.3035832898 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 18 |
Hb_000538_010 |
0.3057702925 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_012573_080 |
0.3059722792 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 20 |
Hb_000465_480 |
0.3085709771 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16835, mitochondrial [Jatropha curcas] |