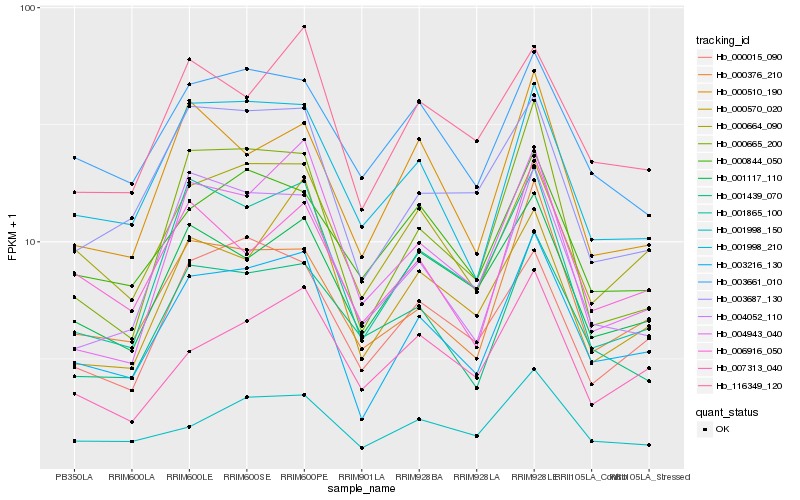
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003216_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 2 |
Hb_000664_090 |
0.0964605144 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 3 |
Hb_001998_210 |
0.0970676506 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_000376_210 |
0.099983065 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 5 |
Hb_001865_100 |
0.1008297503 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 6 |
Hb_003687_130 |
0.1029861975 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 7 |
Hb_000665_200 |
0.1079132463 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 8 |
Hb_116349_120 |
0.1093398583 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_004052_110 |
0.11194517 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000015_090 |
0.1121790236 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 11 |
Hb_000510_190 |
0.1138152212 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 12 |
Hb_001117_110 |
0.1141210266 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 13 |
Hb_001998_150 |
0.1167100772 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 14 |
Hb_006916_050 |
0.1174819727 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 15 |
Hb_001439_070 |
0.1194041982 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000844_050 |
0.1200952263 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 17 |
Hb_007313_040 |
0.1208696207 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 18 |
Hb_003661_010 |
0.1216992453 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004943_040 |
0.1221311068 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000570_020 |
0.1223712072 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |