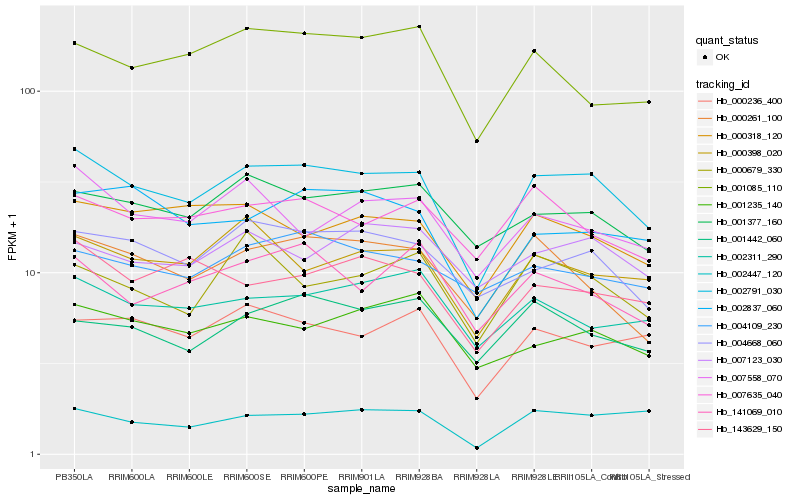
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002791_030 |
0.0 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 2 |
Hb_004668_060 |
0.0936432271 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001377_160 |
0.0972944556 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 4 |
Hb_000398_020 |
0.1000262084 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000318_120 |
0.1015208657 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000261_100 |
0.1041781996 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 7 |
Hb_007558_070 |
0.1046714409 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007635_040 |
0.1052536605 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000236_400 |
0.1056849348 |
- |
- |
hypothetical protein JCGZ_01058 [Jatropha curcas] |
| 10 |
Hb_002837_060 |
0.1061306933 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 11 |
Hb_143629_150 |
0.1077508767 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 12 |
Hb_001235_140 |
0.108288759 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 13 |
Hb_002311_290 |
0.1087579236 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 14 |
Hb_002447_120 |
0.1093897808 |
- |
- |
hypothetical protein O988_01801, partial [Pseudogymnoascus pannorum VKM F-3808] |
| 15 |
Hb_004109_230 |
0.1094524423 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_001442_060 |
0.1107186203 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000679_330 |
0.1116945056 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 18 |
Hb_001085_110 |
0.1121428398 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 19 |
Hb_007123_030 |
0.1125129059 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 20 |
Hb_141069_010 |
0.113566617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |