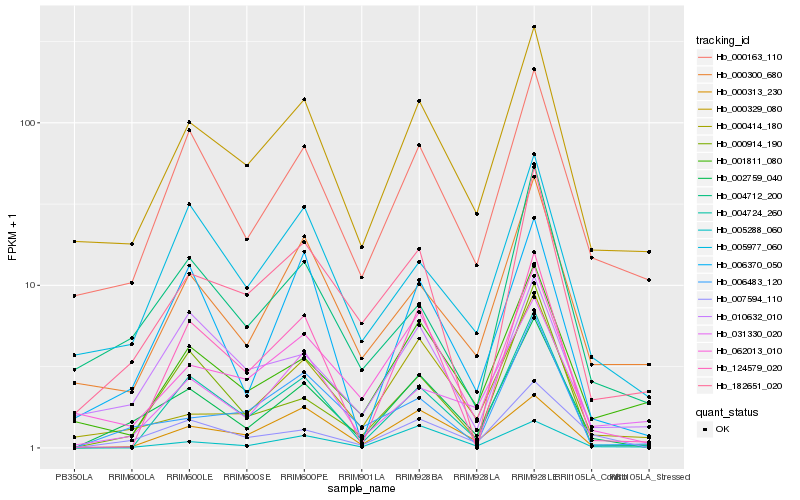
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002759_040 |
0.0 |
- |
- |
BnaA09g29830D [Brassica napus] |
| 2 |
Hb_007594_110 |
0.1374830837 |
- |
- |
fructokinase [Manihot esculenta] |
| 3 |
Hb_000329_080 |
0.1675493684 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000163_110 |
0.1699404773 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 5 |
Hb_004724_260 |
0.1757495947 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 6 |
Hb_006370_050 |
0.1765817158 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_062013_010 |
0.1781386419 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_000914_190 |
0.1837664757 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_182651_020 |
0.1853426216 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000414_180 |
0.1853985219 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 11 |
Hb_004712_200 |
0.1870867152 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 12 |
Hb_006483_120 |
0.1909227375 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_005288_060 |
0.1944635761 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 14 |
Hb_000300_680 |
0.1985278181 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001811_080 |
0.2016843214 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 16 |
Hb_031330_020 |
0.2016957597 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 17 |
Hb_005977_060 |
0.2017136474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010632_010 |
0.2025763337 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_124579_020 |
0.2028062536 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 20 |
Hb_000313_230 |
0.2035149149 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |