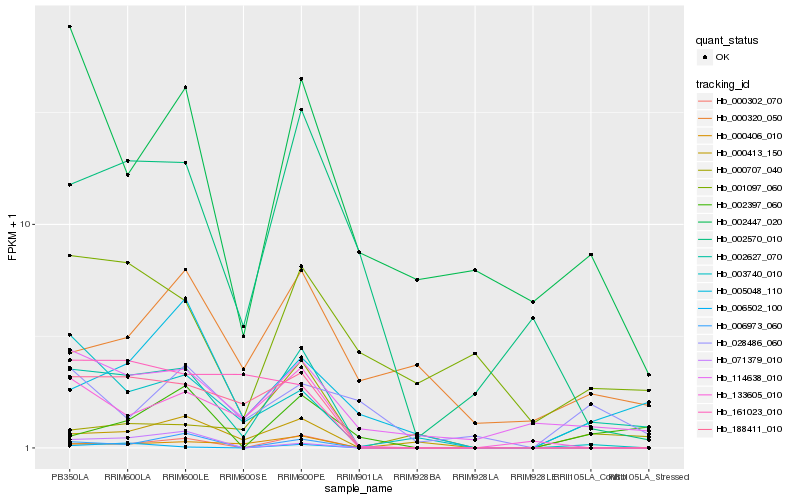
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002627_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_114638_010 |
0.2595056945 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_188411_010 |
0.2777427097 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 [Jatropha curcas] |
| 4 |
Hb_000302_070 |
0.279280521 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 5 |
Hb_133605_010 |
0.2937782511 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_005048_110 |
0.2981541177 |
- |
- |
- |
| 7 |
Hb_002397_060 |
0.3064739329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000406_010 |
0.3072520498 |
- |
- |
PREDICTED: uncharacterized protein LOC105647846 [Jatropha curcas] |
| 9 |
Hb_003740_010 |
0.3080896629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002570_010 |
0.3081321772 |
- |
- |
hypothetical protein JCGZ_25329 [Jatropha curcas] |
| 11 |
Hb_000413_150 |
0.3107761216 |
- |
- |
PREDICTED: uncharacterized protein LOC103709996 [Phoenix dactylifera] |
| 12 |
Hb_006973_060 |
0.3120698489 |
- |
- |
Uncharacterized protein TCM_038475 [Theobroma cacao] |
| 13 |
Hb_002447_020 |
0.3153989241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001097_060 |
0.3162143605 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 8 [Jatropha curcas] |
| 15 |
Hb_000707_040 |
0.3325137745 |
- |
- |
Uncharacterized protein TCM_021829 [Theobroma cacao] |
| 16 |
Hb_006502_100 |
0.3338914989 |
- |
- |
hypothetical protein POPTR_0003s01580g [Populus trichocarpa] |
| 17 |
Hb_000320_050 |
0.3374250989 |
- |
- |
bcr-associated protein, bap, putative [Ricinus communis] |
| 18 |
Hb_071379_010 |
0.3408813336 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 3-like [Nelumbo nucifera] |
| 19 |
Hb_028486_060 |
0.349569856 |
- |
- |
PREDICTED: putative germin-like protein 9-2 [Jatropha curcas] |
| 20 |
Hb_161023_010 |
0.3505803802 |
- |
- |
PREDICTED: uncharacterized protein LOC105639929 [Jatropha curcas] |