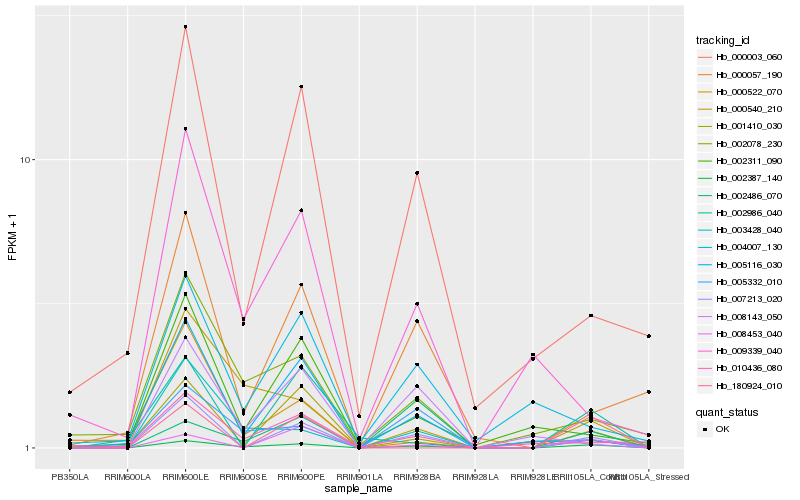
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002387_140 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like [Jatropha curcas] |
| 2 |
Hb_007213_020 |
0.218766105 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000540_210 |
0.2620519228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005116_030 |
0.2690147125 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 5 |
Hb_008453_040 |
0.2798408135 |
- |
- |
PREDICTED: expansin-A11 [Jatropha curcas] |
| 6 |
Hb_002078_230 |
0.2875562837 |
- |
- |
PREDICTED: HORMA domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_000003_060 |
0.2974029751 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 8 |
Hb_000057_190 |
0.2987208182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010436_080 |
0.3013703086 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 10 |
Hb_005332_010 |
0.3032727426 |
- |
- |
hypothetical protein EUGRSUZ_J02276 [Eucalyptus grandis] |
| 11 |
Hb_002486_070 |
0.3048494015 |
transcription factor |
TF Family: LOB |
hypothetical protein POPTR_0008s07280g [Populus trichocarpa] |
| 12 |
Hb_008143_050 |
0.3049025381 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 13 |
Hb_003428_040 |
0.3059483489 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_004007_130 |
0.3123523789 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 15 |
Hb_002311_090 |
0.3124320049 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 16 |
Hb_000522_070 |
0.3134350166 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |
| 17 |
Hb_180924_010 |
0.3144484226 |
- |
- |
CPI-4 [Morus alba var. atropurpurea] |
| 18 |
Hb_001410_030 |
0.3175261481 |
- |
- |
hypothetical protein JCGZ_20419 [Jatropha curcas] |
| 19 |
Hb_009339_040 |
0.3200406925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002986_040 |
0.3207140009 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |