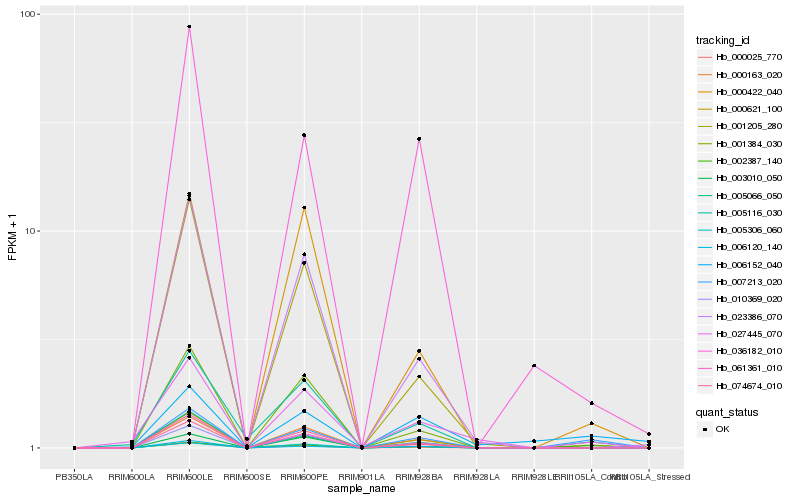
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007213_020 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005116_030 |
0.2132378936 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002387_140 |
0.218766105 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like [Jatropha curcas] |
| 4 |
Hb_027445_070 |
0.2228569607 |
- |
- |
PREDICTED: denticleless protein homolog [Jatropha curcas] |
| 5 |
Hb_036182_010 |
0.2230693234 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 6 |
Hb_010369_020 |
0.2267421729 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 7 |
Hb_000621_100 |
0.2297046536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000163_020 |
0.2316677999 |
- |
- |
- |
| 9 |
Hb_000422_040 |
0.2335222991 |
- |
- |
hypothetical protein CICLE_v10013147mg [Citrus clementina] |
| 10 |
Hb_061361_010 |
0.2339340385 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_000025_770 |
0.2345595935 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 12 |
Hb_006152_040 |
0.2368706062 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105645062 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001205_280 |
0.2405924016 |
- |
- |
PREDICTED: probable polyol transporter 6 [Jatropha curcas] |
| 14 |
Hb_023386_070 |
0.2438560024 |
- |
- |
PREDICTED: probable pectinesterase 68 [Jatropha curcas] |
| 15 |
Hb_005306_060 |
0.2446068797 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 16 |
Hb_006120_140 |
0.2457181824 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 17 |
Hb_074674_010 |
0.2464977165 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 18 |
Hb_005066_050 |
0.2472334139 |
- |
- |
- |
| 19 |
Hb_003010_050 |
0.2491427414 |
- |
- |
- |
| 20 |
Hb_001384_030 |
0.2511128262 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |