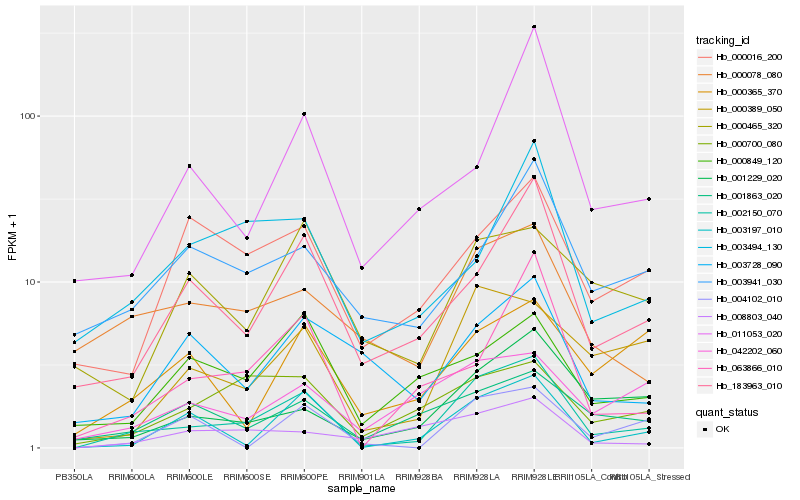
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_070 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 2 |
Hb_183963_010 |
0.2169896557 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 3 |
Hb_003197_010 |
0.2187511696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000389_050 |
0.227230694 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 5 |
Hb_001229_020 |
0.2313273064 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_063866_010 |
0.235748323 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 7 |
Hb_000078_080 |
0.2450193481 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein JCGZ_17040 [Jatropha curcas] |
| 8 |
Hb_000700_080 |
0.2491062164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003494_130 |
0.2570859249 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 10 |
Hb_001863_020 |
0.2592388905 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004102_010 |
0.2607387351 |
- |
- |
F-box family protein [Theobroma cacao] |
| 12 |
Hb_011053_020 |
0.2608029822 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 13 |
Hb_000016_200 |
0.2629713532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003941_030 |
0.2630812559 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 15 |
Hb_000849_120 |
0.2643100028 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 16 |
Hb_042202_060 |
0.2649608335 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Jatropha curcas] |
| 17 |
Hb_008803_040 |
0.2664699155 |
- |
- |
PREDICTED: uncharacterized protein LOC105157106 [Sesamum indicum] |
| 18 |
Hb_003728_090 |
0.2678013677 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 19 |
Hb_000465_320 |
0.2724747901 |
- |
- |
PREDICTED: nudix hydrolase 8 [Jatropha curcas] |
| 20 |
Hb_000365_370 |
0.2733347139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |