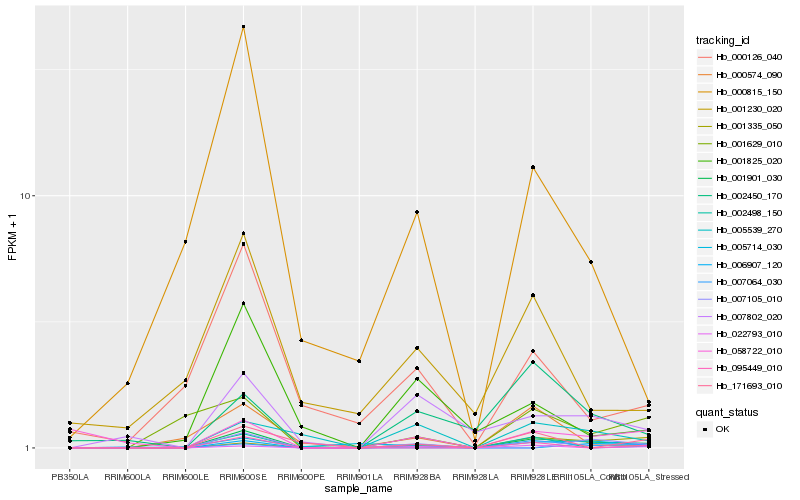
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001901_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_007802_020 |
0.3144112041 |
- |
- |
PREDICTED: receptor-like protein 12 [Eucalyptus grandis] |
| 3 |
Hb_001335_050 |
0.3220373211 |
- |
- |
- |
| 4 |
Hb_005539_270 |
0.3227051356 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_002498_150 |
0.327003911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001629_010 |
0.3357944669 |
- |
- |
Disease resistance protein RPM1, putative [Theobroma cacao] |
| 7 |
Hb_002450_170 |
0.3412391386 |
transcription factor |
TF Family: FAR1 |
Protein FAR1-RELATED SEQUENCE 5 [Glycine soja] |
| 8 |
Hb_005714_030 |
0.3442205568 |
- |
- |
PREDICTED: COBRA-like protein 6 isoform X1 [Gossypium raimondii] |
| 9 |
Hb_022793_010 |
0.3452315088 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 10 |
Hb_171693_010 |
0.3502209038 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 11 |
Hb_000815_150 |
0.3504421466 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_095449_010 |
0.3513441084 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein-like [Jatropha curcas] |
| 13 |
Hb_001825_020 |
0.3523083298 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_007064_030 |
0.3562929642 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 15 |
Hb_007105_010 |
0.3618163032 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |
| 16 |
Hb_000126_040 |
0.3629952174 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_058722_010 |
0.3664426735 |
- |
- |
- |
| 18 |
Hb_000574_090 |
0.3672348704 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 19 |
Hb_006907_120 |
0.3698735864 |
- |
- |
PREDICTED: uncharacterized protein LOC103327233 [Prunus mume] |
| 20 |
Hb_001230_020 |
0.3710291951 |
- |
- |
hypothetical protein POPTR_0001s31610g [Populus trichocarpa] |