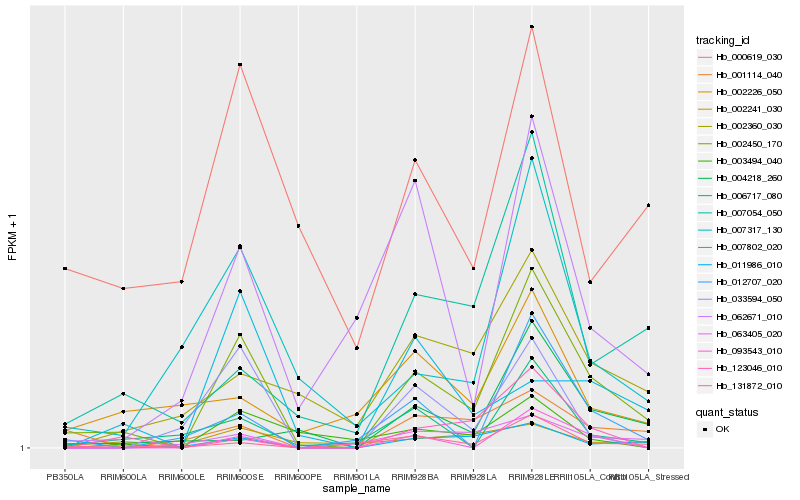
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002450_170 |
0.0 |
transcription factor |
TF Family: FAR1 |
Protein FAR1-RELATED SEQUENCE 5 [Glycine soja] |
| 2 |
Hb_007054_050 |
0.2739904979 |
- |
- |
- |
| 3 |
Hb_131872_010 |
0.2778041743 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_001114_040 |
0.2804822976 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 5 |
Hb_011986_010 |
0.2868726832 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_007317_130 |
0.2936870046 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 7 |
Hb_063405_020 |
0.297777281 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 8 |
Hb_093543_010 |
0.3012653853 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 9 |
Hb_002360_030 |
0.3054047595 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 10 |
Hb_006717_080 |
0.30554786 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 11 |
Hb_007802_020 |
0.3061635142 |
- |
- |
PREDICTED: receptor-like protein 12 [Eucalyptus grandis] |
| 12 |
Hb_062671_010 |
0.3071965909 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002241_030 |
0.3087449158 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 14 |
Hb_000619_030 |
0.3193804343 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_004218_260 |
0.3198495988 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 16 |
Hb_123046_010 |
0.3207079499 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 17 |
Hb_002226_050 |
0.3212038017 |
- |
- |
PREDICTED: uncharacterized protein LOC105641366 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012707_020 |
0.3218099436 |
- |
- |
- |
| 19 |
Hb_033594_050 |
0.324261527 |
- |
- |
PREDICTED: cyclin-D4-1-like [Jatropha curcas] |
| 20 |
Hb_003494_040 |
0.324825306 |
transcription factor |
TF Family: NAC |
PREDICTED: protein FEZ [Jatropha curcas] |