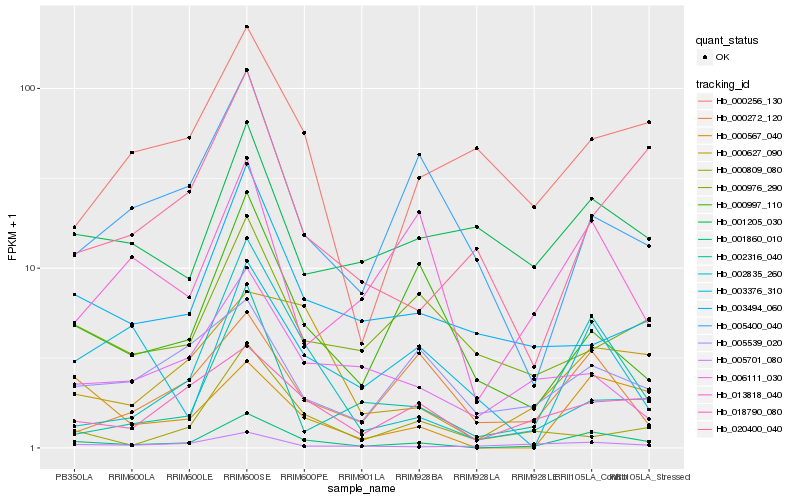
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001860_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_018790_080 |
0.2287114186 |
- |
- |
hypothetical protein POPTR_0011s06335g [Populus trichocarpa] |
| 3 |
Hb_000627_090 |
0.2380817238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002316_040 |
0.2399761862 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 5 |
Hb_000567_040 |
0.2415509963 |
- |
- |
PREDICTED: uncharacterized protein LOC105649657 [Jatropha curcas] |
| 6 |
Hb_005701_080 |
0.2498817824 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000997_110 |
0.2567387515 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 8 |
Hb_000272_120 |
0.2637928133 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR18 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003494_060 |
0.2654206285 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006111_030 |
0.2667461607 |
- |
- |
PREDICTED: phospholipase A(1) LCAT3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_013818_040 |
0.2677435046 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 12 |
Hb_005539_020 |
0.2696556041 |
- |
- |
- |
| 13 |
Hb_000809_080 |
0.2697724581 |
- |
- |
50S ribosomal protein L25, putative [Ricinus communis] |
| 14 |
Hb_001205_030 |
0.2697787386 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0007s12970g [Populus trichocarpa] |
| 15 |
Hb_000976_290 |
0.2722130522 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 16 |
Hb_005400_040 |
0.2732772623 |
- |
- |
PREDICTED: uncharacterized protein At4g04775-like [Jatropha curcas] |
| 17 |
Hb_000256_130 |
0.2737994942 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 18 |
Hb_002835_260 |
0.2757474957 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 19 |
Hb_020400_040 |
0.276182875 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 20 |
Hb_003376_310 |
0.2762533153 |
- |
- |
hypothetical protein JCGZ_16500 [Jatropha curcas] |