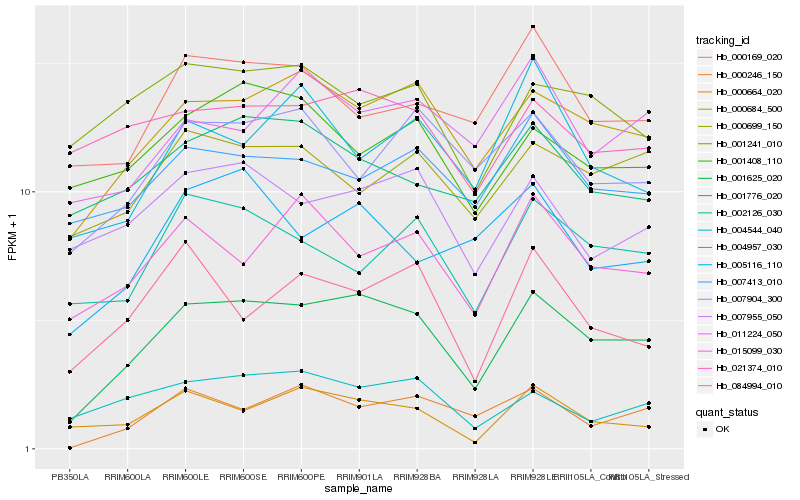
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001625_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 2 |
Hb_000684_500 |
0.0995735421 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_015099_030 |
0.1337117613 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 4 |
Hb_007955_050 |
0.1390677469 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_002126_030 |
0.1412825124 |
- |
- |
ferrochelatase, putative [Ricinus communis] |
| 6 |
Hb_004544_040 |
0.1419575956 |
- |
- |
hypothetical protein PHAVU_010G103000g [Phaseolus vulgaris] |
| 7 |
Hb_007904_300 |
0.1426243913 |
- |
- |
copine, putative [Ricinus communis] |
| 8 |
Hb_005116_110 |
0.1427272256 |
- |
- |
PREDICTED: uncharacterized protein LOC105640215 [Jatropha curcas] |
| 9 |
Hb_084994_010 |
0.143314694 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007413_010 |
0.1439394576 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 11 |
Hb_000699_150 |
0.1444301436 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 12 |
Hb_000246_150 |
0.1444332614 |
- |
- |
PREDICTED: protein FAM135B [Jatropha curcas] |
| 13 |
Hb_000664_020 |
0.1450799169 |
- |
- |
tryptophanyl-tRNA synthetase, putative [Ricinus communis] |
| 14 |
Hb_001776_020 |
0.1452175733 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 15 |
Hb_011224_050 |
0.1460976719 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Populus euphratica] |
| 16 |
Hb_000169_020 |
0.147615737 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 17 |
Hb_004957_030 |
0.1484039737 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 18 |
Hb_001408_110 |
0.1498529375 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 19 |
Hb_001241_010 |
0.1507802428 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_021374_010 |
0.1509580063 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |