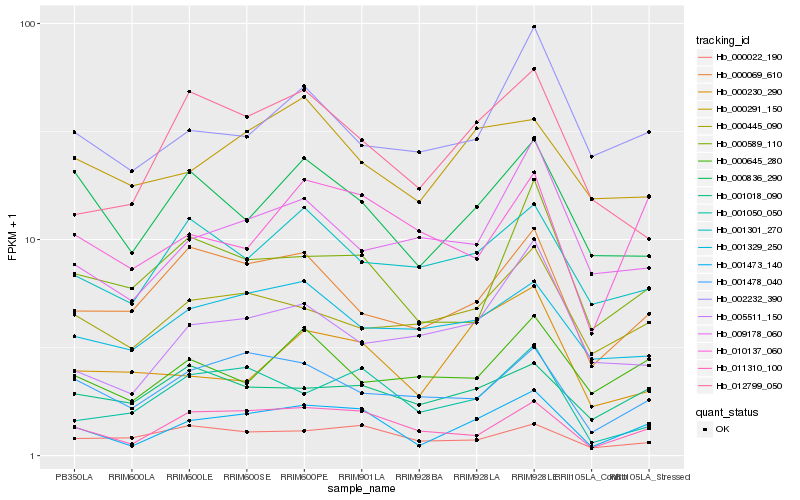
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001473_140 |
0.0 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 2 |
Hb_011310_100 |
0.1300990109 |
- |
- |
hypothetical protein CICLE_v10008199mg [Citrus clementina] |
| 3 |
Hb_001478_040 |
0.1675555605 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |
| 4 |
Hb_000836_290 |
0.1684833459 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000069_610 |
0.1698146375 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 6 |
Hb_000589_110 |
0.1729176587 |
- |
- |
PREDICTED: uncharacterized protein LOC105647614 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001050_050 |
0.1753649649 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g57250, mitochondrial [Jatropha curcas] |
| 8 |
Hb_010137_060 |
0.1758750377 |
- |
- |
PREDICTED: uncharacterized protein LOC105634409 [Jatropha curcas] |
| 9 |
Hb_000445_090 |
0.1761690367 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 10 |
Hb_000230_290 |
0.1771070464 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 11 |
Hb_001301_270 |
0.177816584 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 12 |
Hb_000022_190 |
0.1788091576 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |
| 13 |
Hb_012799_050 |
0.1801713261 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 14 |
Hb_000291_150 |
0.1801887392 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 15 |
Hb_001018_090 |
0.1802001904 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 16 |
Hb_009178_060 |
0.1811575104 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_002232_390 |
0.1815126695 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 18 |
Hb_001329_250 |
0.1822827778 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000645_280 |
0.1846066891 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 20 |
Hb_005511_150 |
0.1848987096 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic isoform X5 [Populus euphratica] |