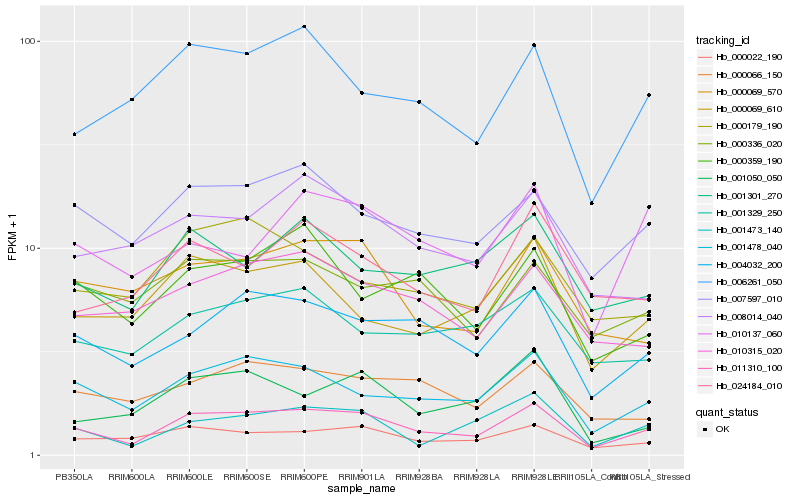
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011310_100 |
0.0 |
- |
- |
hypothetical protein CICLE_v10008199mg [Citrus clementina] |
| 2 |
Hb_001478_040 |
0.1066193743 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |
| 3 |
Hb_004032_200 |
0.1096803984 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 4 |
Hb_000022_190 |
0.1212166637 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |
| 5 |
Hb_007597_010 |
0.1262210839 |
- |
- |
hypothetical protein B456_010G072300 [Gossypium raimondii] |
| 6 |
Hb_006261_050 |
0.129874643 |
transcription factor |
TF Family: C2C2-YABBY |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 7 |
Hb_001473_140 |
0.1300990109 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 8 |
Hb_000069_610 |
0.135686463 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 9 |
Hb_000066_150 |
0.1364552159 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 10 |
Hb_010315_020 |
0.1377631153 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 11 |
Hb_000069_570 |
0.1381655993 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_008014_040 |
0.1390226114 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 13 |
Hb_000359_190 |
0.1404586273 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Nicotiana sylvestris] |
| 14 |
Hb_001301_270 |
0.1416531952 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 15 |
Hb_001329_250 |
0.1422020195 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000336_020 |
0.1439193974 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 17 |
Hb_010137_060 |
0.144131705 |
- |
- |
PREDICTED: uncharacterized protein LOC105634409 [Jatropha curcas] |
| 18 |
Hb_024184_010 |
0.1446577119 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_000179_190 |
0.1449688622 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 20 |
Hb_001050_050 |
0.1455465255 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g57250, mitochondrial [Jatropha curcas] |