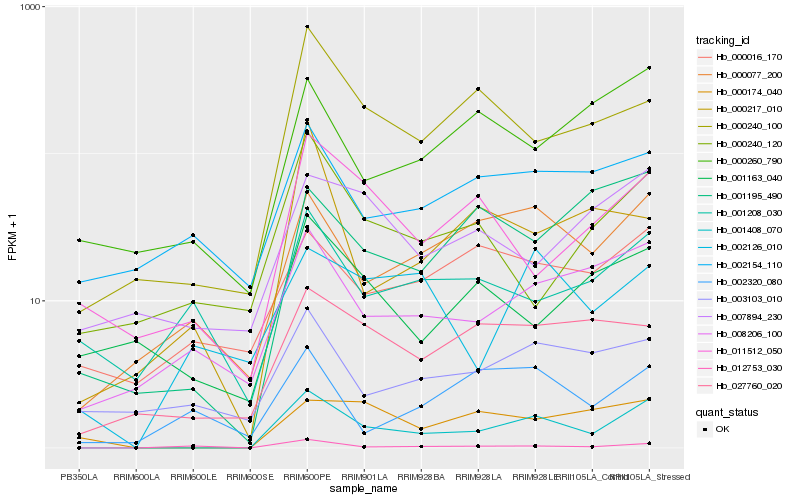
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001408_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000077_200 |
0.2269416217 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 3 |
Hb_008206_100 |
0.2384940286 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 4 |
Hb_000240_100 |
0.2399083667 |
- |
- |
PREDICTED: ras-related protein Rab5-like [Eucalyptus grandis] |
| 5 |
Hb_012753_030 |
0.2426905706 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 6 |
Hb_001195_490 |
0.2574566537 |
- |
- |
hypothetical protein glysoja_018593 [Glycine soja] |
| 7 |
Hb_000174_040 |
0.2615186037 |
- |
- |
- |
| 8 |
Hb_027760_020 |
0.2636789817 |
- |
- |
hypothetical protein M569_04462, partial [Genlisea aurea] |
| 9 |
Hb_011512_050 |
0.2691493216 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 10 |
Hb_000260_790 |
0.2717017277 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 12 isoform 1 [Theobroma cacao] |
| 11 |
Hb_000016_170 |
0.2770438263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003103_010 |
0.2804057744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002154_110 |
0.283743065 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 14 |
Hb_007894_230 |
0.2841539727 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 15 |
Hb_001208_030 |
0.2844427159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000240_120 |
0.2850067138 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 17 |
Hb_001163_040 |
0.2872574031 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 18 |
Hb_002126_010 |
0.2874804712 |
- |
- |
predicted protein [Hordeum vulgare subsp. vulgare] |
| 19 |
Hb_000217_010 |
0.2879042303 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 20 |
Hb_002320_080 |
0.2884518633 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |