| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
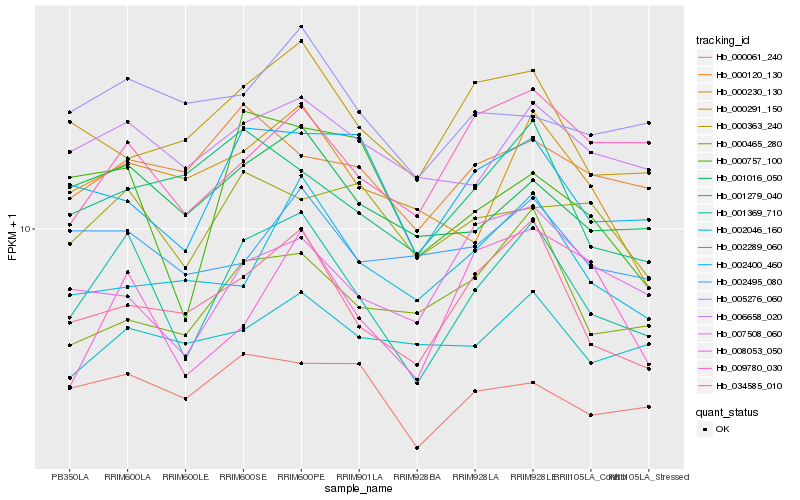
Hb_001369_710 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648579 isoform X1 [Jatropha curcas] |
| 2 |
Hb_034585_010 |
0.1206754736 |
- |
- |
PREDICTED: uncharacterized protein LOC105639637 [Jatropha curcas] |
| 3 |
Hb_001016_050 |
0.1309582583 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 4 |
Hb_006658_020 |
0.1347828297 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 5 |
Hb_002400_460 |
0.136599216 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000230_130 |
0.1425879534 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000061_240 |
0.1435593004 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_000757_100 |
0.1446639586 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 2 [Theobroma cacao] |
| 9 |
Hb_000291_150 |
0.14810007 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 10 |
Hb_009780_030 |
0.1483457304 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 11 |
Hb_000465_280 |
0.1489784087 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 12 |
Hb_001279_040 |
0.1503374084 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 13 |
Hb_000363_240 |
0.1519042743 |
- |
- |
PREDICTED: beta-(1,2)-xylosyltransferase [Jatropha curcas] |
| 14 |
Hb_002046_160 |
0.1525958964 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_005276_060 |
0.1532998698 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002495_080 |
0.1543020549 |
- |
- |
PREDICTED: polygalacturonase ADPG2 [Jatropha curcas] |
| 17 |
Hb_000120_130 |
0.154501787 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 18 |
Hb_007508_060 |
0.1561595915 |
- |
- |
PREDICTED: uncharacterized protein LOC105640628 [Jatropha curcas] |
| 19 |
Hb_002289_060 |
0.1563230643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008053_050 |
0.1571727275 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |