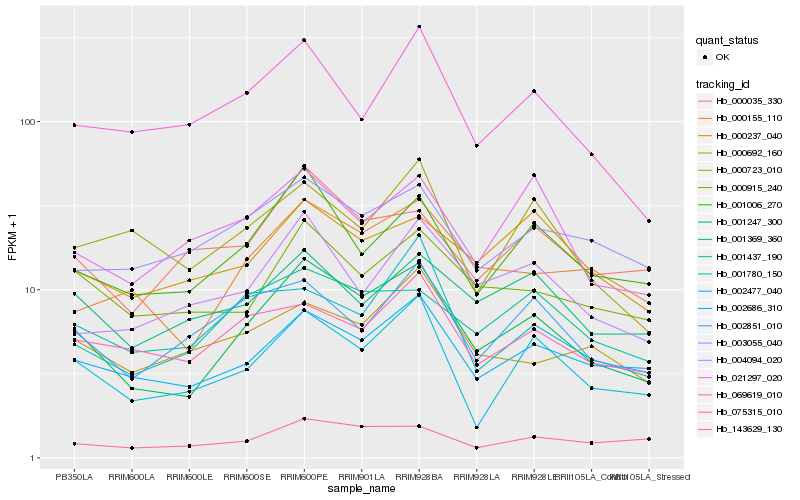
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_300 |
0.0 |
- |
- |
hypothetical protein JCGZ_23167 [Jatropha curcas] |
| 2 |
Hb_001006_270 |
0.1279671986 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 3 |
Hb_000155_110 |
0.1296712437 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 4 |
Hb_002686_310 |
0.1304743883 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 5 |
Hb_002477_040 |
0.1311628024 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein isoform 2 [Theobroma cacao] |
| 6 |
Hb_000915_240 |
0.1426853731 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_001437_190 |
0.1492507743 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_004094_020 |
0.1505116883 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_069619_010 |
0.151982399 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 10 |
Hb_002851_010 |
0.1534219198 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_143629_130 |
0.1535229741 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000035_330 |
0.1535523251 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |
| 13 |
Hb_001369_360 |
0.1544400783 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 14 |
Hb_003055_040 |
0.1566587165 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000237_040 |
0.1568682492 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 16 |
Hb_075315_010 |
0.1577344931 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 17 |
Hb_021297_020 |
0.15992807 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 18 |
Hb_000692_160 |
0.1651369435 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 19 |
Hb_001780_150 |
0.1652877225 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 20 |
Hb_000723_010 |
0.165882516 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |