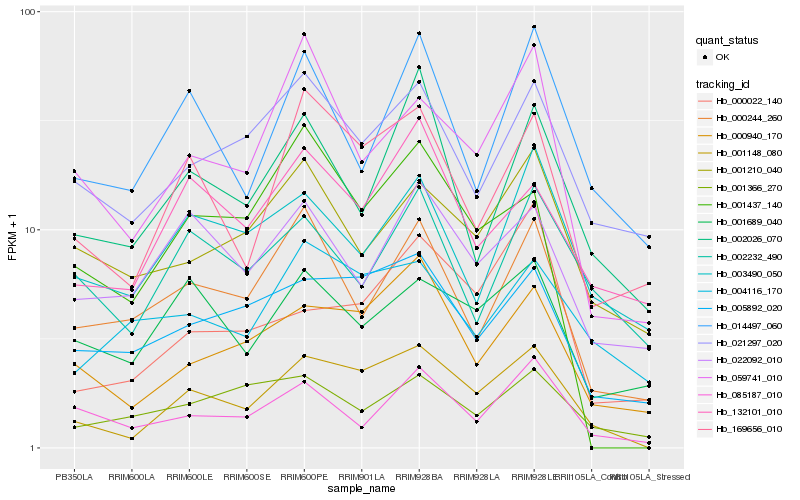
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001148_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 2 |
Hb_000940_170 |
0.1531121791 |
- |
- |
hypothetical protein VITISV_029834 [Vitis vinifera] |
| 3 |
Hb_169656_010 |
0.1594307698 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 4 |
Hb_005892_020 |
0.1716538662 |
- |
- |
PREDICTED: uncharacterized protein LOC105632170 isoform X1 [Jatropha curcas] |
| 5 |
Hb_059741_010 |
0.1880504645 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 6 |
Hb_132101_010 |
0.1887238961 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 7 |
Hb_021297_020 |
0.1914712005 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 8 |
Hb_001437_140 |
0.1926834262 |
- |
- |
- |
| 9 |
Hb_001689_040 |
0.19775395 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 1-like [Jatropha curcas] |
| 10 |
Hb_000022_140 |
0.1979828981 |
- |
- |
hypothetical protein MIMGU_mgv1a009920mg [Erythranthe guttata] |
| 11 |
Hb_002026_070 |
0.1981055565 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 12 |
Hb_004116_170 |
0.1990895651 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000244_260 |
0.1996536804 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 14 |
Hb_014497_060 |
0.2011655133 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 15 |
Hb_022092_010 |
0.2036860743 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 16 |
Hb_003490_050 |
0.2049270384 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001210_040 |
0.2054313993 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 18 |
Hb_085187_010 |
0.2108683106 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 19 |
Hb_002232_490 |
0.2113533651 |
- |
- |
PREDICTED: probable sphingolipid transporter spinster homolog 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001366_270 |
0.2119742093 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |