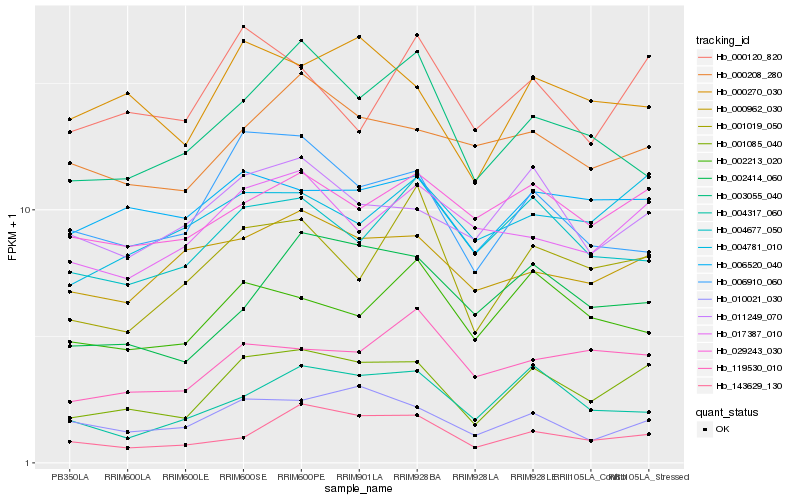
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 2 |
Hb_000270_030 |
0.1233271471 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0015s07640g [Populus trichocarpa] |
| 3 |
Hb_010021_030 |
0.1244665955 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 4 |
Hb_002414_060 |
0.12656756 |
- |
- |
hypothetical protein EUGRSUZ_E01310 [Eucalyptus grandis] |
| 5 |
Hb_011249_070 |
0.1277289618 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 6 |
Hb_001019_050 |
0.1283290698 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 7 |
Hb_143629_130 |
0.1286912235 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_029243_030 |
0.1303149102 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004781_010 |
0.1306303224 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 10 |
Hb_006910_060 |
0.1319229586 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 11 |
Hb_004317_060 |
0.1328182781 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_017387_010 |
0.134051585 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 13 |
Hb_000208_280 |
0.1345653701 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 14 |
Hb_000962_030 |
0.1346315398 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 15 |
Hb_006520_040 |
0.1349374377 |
- |
- |
PREDICTED: uncharacterized protein LOC105641714 isoform X2 [Jatropha curcas] |
| 16 |
Hb_119530_010 |
0.1351313354 |
- |
- |
- |
| 17 |
Hb_004677_050 |
0.1393761028 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000120_820 |
0.1401739924 |
- |
- |
dihydrodipicolinate synthase, putative [Ricinus communis] |
| 19 |
Hb_003055_040 |
0.1415817477 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002213_020 |
0.1420587103 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |