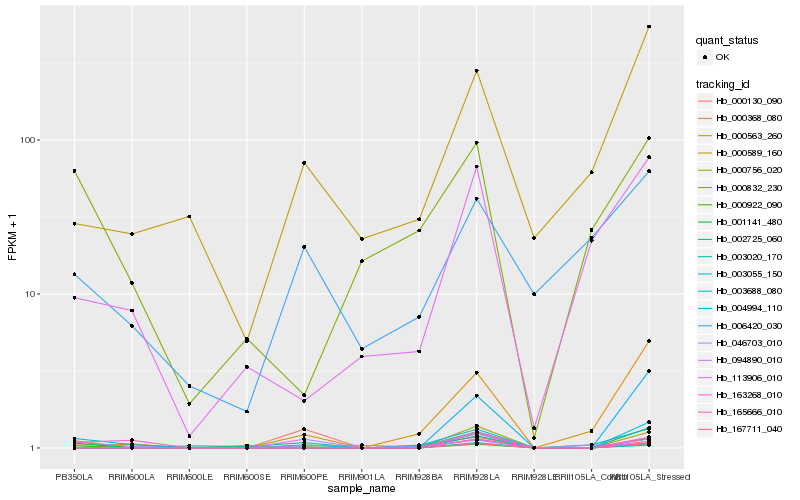
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_090 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g11290-like [Populus euphratica] |
| 2 |
Hb_001141_480 |
0.2404027796 |
- |
- |
- |
| 3 |
Hb_003688_080 |
0.3045582688 |
- |
- |
- |
| 4 |
Hb_094890_010 |
0.3803184183 |
- |
- |
- |
| 5 |
Hb_165666_010 |
0.3861438801 |
- |
- |
- |
| 6 |
Hb_000589_160 |
0.3931001559 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 7 |
Hb_000832_230 |
0.3977727082 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |
| 8 |
Hb_167711_040 |
0.3996064176 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |
| 9 |
Hb_000368_080 |
0.4013135378 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 10 |
Hb_000756_020 |
0.408685297 |
- |
- |
BnaA05g19440D [Brassica napus] |
| 11 |
Hb_006420_030 |
0.4088858574 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 12 |
Hb_113906_010 |
0.4091777929 |
- |
- |
hypothetical protein JCGZ_18857 [Jatropha curcas] |
| 13 |
Hb_000130_090 |
0.4092541052 |
- |
- |
Anther-specific protein LAT52 precursor, putative [Ricinus communis] |
| 14 |
Hb_000563_260 |
0.4111675106 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 15 |
Hb_002725_060 |
0.4114373718 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 16 |
Hb_004994_110 |
0.4121201709 |
- |
- |
- |
| 17 |
Hb_003020_170 |
0.4158879122 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 18 |
Hb_046703_010 |
0.4161030965 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 19 |
Hb_003055_150 |
0.4164110423 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 20 |
Hb_163268_010 |
0.4171985369 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |