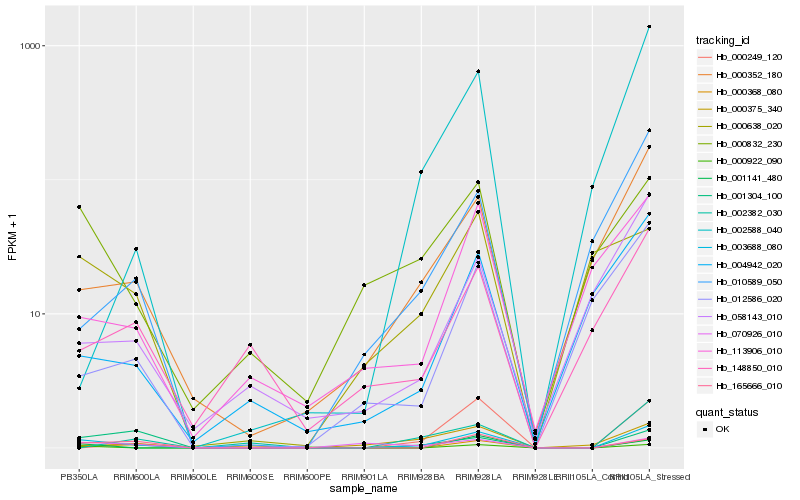
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_001141_480 |
0.2659024749 |
- |
- |
- |
| 3 |
Hb_165666_010 |
0.2701581635 |
- |
- |
- |
| 4 |
Hb_000352_180 |
0.2741354715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000368_080 |
0.2838525918 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 6 |
Hb_000832_230 |
0.2963768676 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |
| 7 |
Hb_000249_120 |
0.2991600998 |
- |
- |
PREDICTED: uncharacterized protein LOC104613364 [Nelumbo nucifera] |
| 8 |
Hb_070926_010 |
0.3010455807 |
- |
- |
PREDICTED: mRNA-decapping enzyme subunit 2-like [Populus euphratica] |
| 9 |
Hb_000922_090 |
0.3045582688 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g11290-like [Populus euphratica] |
| 10 |
Hb_001304_100 |
0.3099025793 |
- |
- |
- |
| 11 |
Hb_113906_010 |
0.3166239225 |
- |
- |
hypothetical protein JCGZ_18857 [Jatropha curcas] |
| 12 |
Hb_000375_340 |
0.3175422875 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Populus euphratica] |
| 13 |
Hb_148850_010 |
0.3225492959 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 14 |
Hb_010589_050 |
0.3247040929 |
- |
- |
- |
| 15 |
Hb_004942_020 |
0.3247533082 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 16 |
Hb_002588_040 |
0.328195881 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 17 |
Hb_058143_010 |
0.32949809 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 18 |
Hb_002382_030 |
0.3363616254 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Malus domestica] |
| 19 |
Hb_012586_020 |
0.3365623928 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000638_020 |
0.3377700186 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 9-like [Jatropha curcas] |