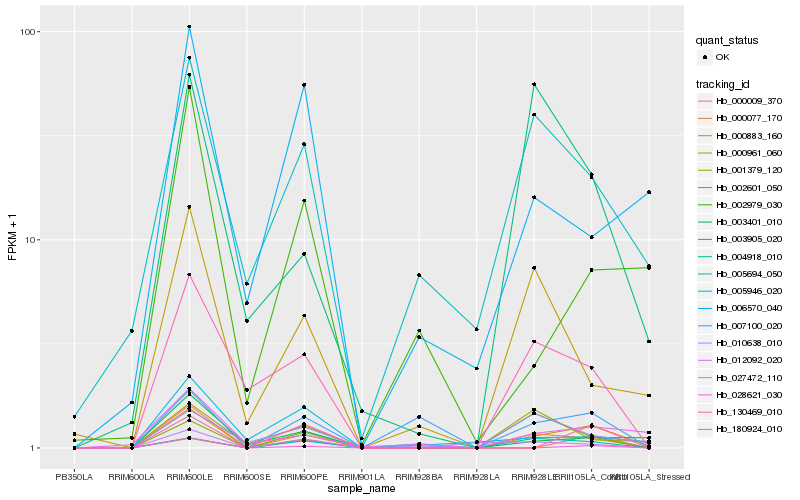
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000883_160 |
0.0 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_130469_010 |
0.2605805302 |
- |
- |
pleiotropic drug resistance ABC transporter family protein, partial [Eleutherococcus senticosus] |
| 3 |
Hb_003401_010 |
0.2694662885 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 4 |
Hb_027472_110 |
0.27134536 |
- |
- |
Patellin-4, putative [Ricinus communis] |
| 5 |
Hb_005694_050 |
0.2835229408 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_000077_170 |
0.2926529933 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Jatropha curcas] |
| 7 |
Hb_003905_020 |
0.300019457 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 8 |
Hb_010638_010 |
0.3027054915 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 9 |
Hb_000961_060 |
0.3037764348 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 10 |
Hb_000009_370 |
0.3050510466 |
- |
- |
hypothetical protein JCGZ_12284 [Jatropha curcas] |
| 11 |
Hb_180924_010 |
0.3057695466 |
- |
- |
CPI-4 [Morus alba var. atropurpurea] |
| 12 |
Hb_028621_030 |
0.3074102522 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 13 |
Hb_001379_120 |
0.3157733802 |
- |
- |
hypothetical protein POPTR_0001s29730g [Populus trichocarpa] |
| 14 |
Hb_007100_020 |
0.3168087623 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 15 |
Hb_002601_050 |
0.3215284584 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 16 |
Hb_012092_020 |
0.3255236624 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 17 |
Hb_006570_040 |
0.3275841085 |
- |
- |
hypothetical protein CICLE_v10002683mg [Citrus clementina] |
| 18 |
Hb_004918_010 |
0.3300683965 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_002979_030 |
0.3308302728 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005946_020 |
0.3316737116 |
- |
- |
Pleiotropic drug resistance protein 4 [Triticum urartu] |