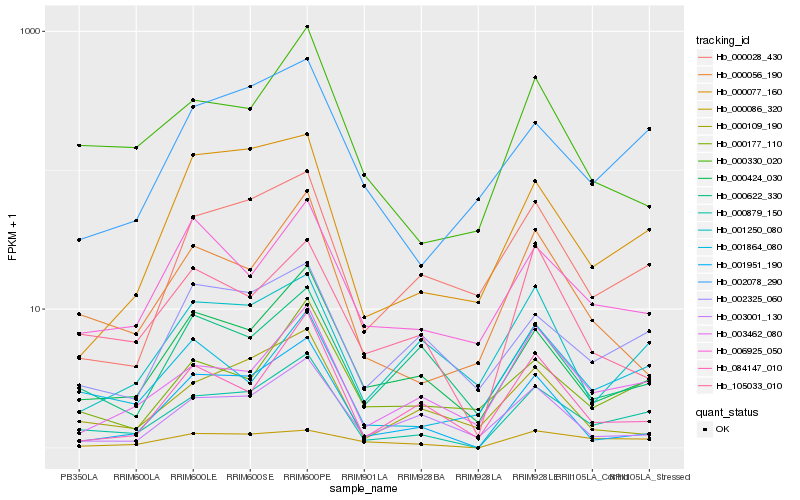
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000879_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000424_030 |
0.1562683473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003462_080 |
0.1631400415 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_001951_190 |
0.1756273623 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 5 |
Hb_002078_290 |
0.1826307173 |
- |
- |
PREDICTED: uncharacterized protein LOC100852520 [Vitis vinifera] |
| 6 |
Hb_000028_430 |
0.1838635591 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000177_110 |
0.1852442644 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 8 |
Hb_000077_160 |
0.1855162583 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 9 |
Hb_001864_080 |
0.1858113162 |
- |
- |
PREDICTED: uncharacterized protein LOC105649405 [Jatropha curcas] |
| 10 |
Hb_000056_190 |
0.1870538029 |
- |
- |
hypothetical protein [Ricinus communis] |
| 11 |
Hb_000330_020 |
0.1873524744 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 12 |
Hb_002325_060 |
0.187689356 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 13 |
Hb_006925_050 |
0.1963832324 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 14 |
Hb_000109_190 |
0.1973941771 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 15 |
Hb_000622_330 |
0.1986059166 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 16 |
Hb_003001_130 |
0.1991576171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_105033_010 |
0.20019092 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_084147_010 |
0.2018166117 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_001250_080 |
0.2023048825 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 20 |
Hb_000086_320 |
0.2034392922 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |