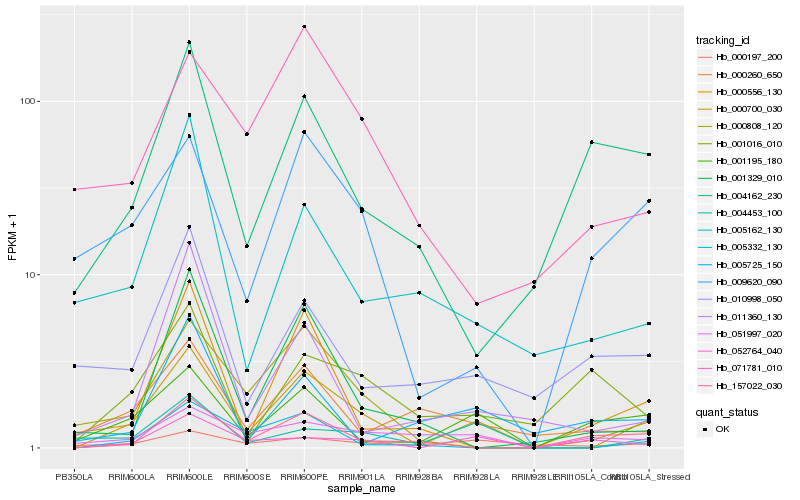
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000700_030 |
0.0 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 2 |
Hb_001195_180 |
0.2373618813 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 3 |
Hb_052764_040 |
0.2499607301 |
- |
- |
syntaxin 125 family protein [Populus trichocarpa] |
| 4 |
Hb_009620_090 |
0.2627215037 |
- |
- |
hypothetical protein L484_000980 [Morus notabilis] |
| 5 |
Hb_000808_120 |
0.2674962157 |
- |
- |
- |
| 6 |
Hb_157022_030 |
0.2675008084 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 7 |
Hb_000260_650 |
0.2713957377 |
- |
- |
PREDICTED: protein PFC0760c [Jatropha curcas] |
| 8 |
Hb_005162_130 |
0.2743131359 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X2 [Jatropha curcas] |
| 9 |
Hb_051997_020 |
0.2762416239 |
- |
- |
PREDICTED: uncharacterized protein LOC100266863 [Vitis vinifera] |
| 10 |
Hb_004453_100 |
0.2765774868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000556_130 |
0.2774856899 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 12 |
Hb_010998_050 |
0.2810916448 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 13 |
Hb_005332_130 |
0.2822411549 |
- |
- |
Mavicyanin, putative [Ricinus communis] |
| 14 |
Hb_001016_010 |
0.282992547 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004162_230 |
0.2838129808 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 16 |
Hb_011360_130 |
0.2850533718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001329_010 |
0.2878935363 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000197_200 |
0.2904202357 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 19 |
Hb_071781_010 |
0.2917044586 |
- |
- |
hypothetical protein POPTR_0010s13190g [Populus trichocarpa] |
| 20 |
Hb_005725_150 |
0.2924448741 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |