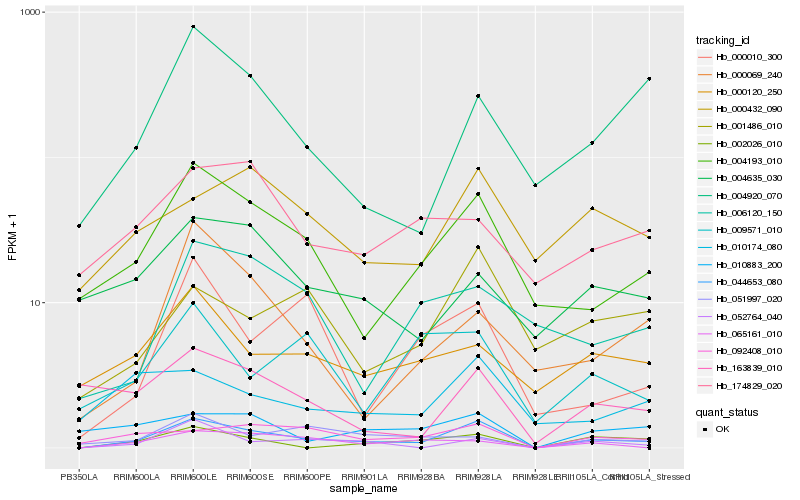
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044653_080 |
0.0 |
- |
- |
Uncharacterized protein TCM_045144 [Theobroma cacao] |
| 2 |
Hb_004193_010 |
0.2196536663 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 3 |
Hb_092408_010 |
0.2442134052 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002026_010 |
0.2567128745 |
transcription factor |
TF Family: mTERF |
hypothetical protein CICLE_v10020375mg [Citrus clementina] |
| 5 |
Hb_052764_040 |
0.2584690267 |
- |
- |
syntaxin 125 family protein [Populus trichocarpa] |
| 6 |
Hb_000432_090 |
0.2637048636 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 7 |
Hb_001486_010 |
0.2654858857 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 8 |
Hb_004920_070 |
0.2712025255 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 9 |
Hb_010174_080 |
0.2735317273 |
- |
- |
- |
| 10 |
Hb_010883_200 |
0.2754950448 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 11 |
Hb_000120_250 |
0.2757509833 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 12 |
Hb_065161_010 |
0.2780056274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000010_300 |
0.2792103468 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_051997_020 |
0.2805326375 |
- |
- |
PREDICTED: uncharacterized protein LOC100266863 [Vitis vinifera] |
| 15 |
Hb_163839_010 |
0.2809882054 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_004635_030 |
0.284069582 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 17 |
Hb_009571_010 |
0.2857564552 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 18 |
Hb_006120_150 |
0.28638168 |
- |
- |
invertase [Hevea brasiliensis] |
| 19 |
Hb_000069_240 |
0.2874748094 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3-like [Jatropha curcas] |
| 20 |
Hb_174829_020 |
0.2887532639 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |