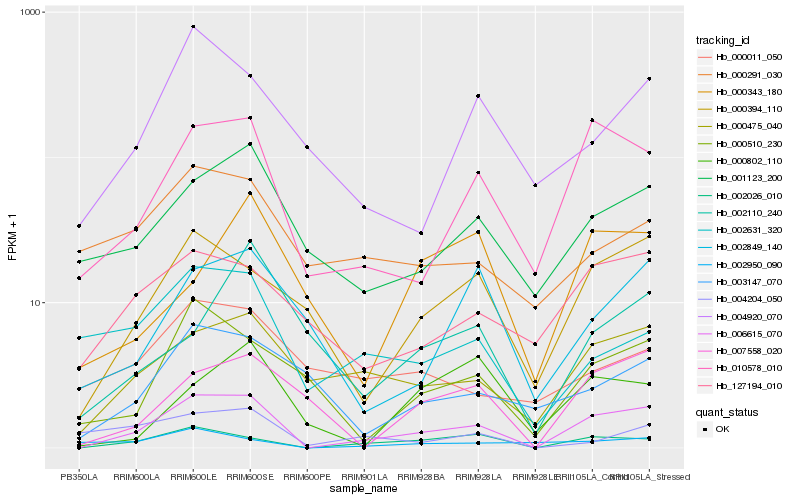
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006615_070 |
0.0 |
- |
- |
hypothetical protein RCOM_1466910 [Ricinus communis] |
| 2 |
Hb_002026_010 |
0.2040248989 |
transcription factor |
TF Family: mTERF |
hypothetical protein CICLE_v10020375mg [Citrus clementina] |
| 3 |
Hb_010578_010 |
0.220113014 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 4 |
Hb_000475_040 |
0.2382515113 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000394_110 |
0.246685425 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 6 |
Hb_001123_200 |
0.2664279837 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007558_020 |
0.2669293368 |
- |
- |
hypothetical protein JCGZ_17631 [Jatropha curcas] |
| 8 |
Hb_000510_230 |
0.2682847497 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 9 |
Hb_127194_010 |
0.2748900276 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 10 |
Hb_004920_070 |
0.2856344215 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 11 |
Hb_002631_320 |
0.2911554195 |
- |
- |
hypothetical protein JCGZ_22628 [Jatropha curcas] |
| 12 |
Hb_002849_140 |
0.2914001186 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 13 |
Hb_003147_070 |
0.2914839985 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 14 |
Hb_004204_050 |
0.3019825225 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000291_030 |
0.3072298035 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 16 |
Hb_000802_110 |
0.3103368805 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 17 |
Hb_002950_090 |
0.3105890756 |
- |
- |
- |
| 18 |
Hb_000011_050 |
0.3108268594 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 19 |
Hb_002110_240 |
0.3131822651 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 20 |
Hb_000343_180 |
0.3139347839 |
- |
- |
NDWp3 [Podospora anserina] |