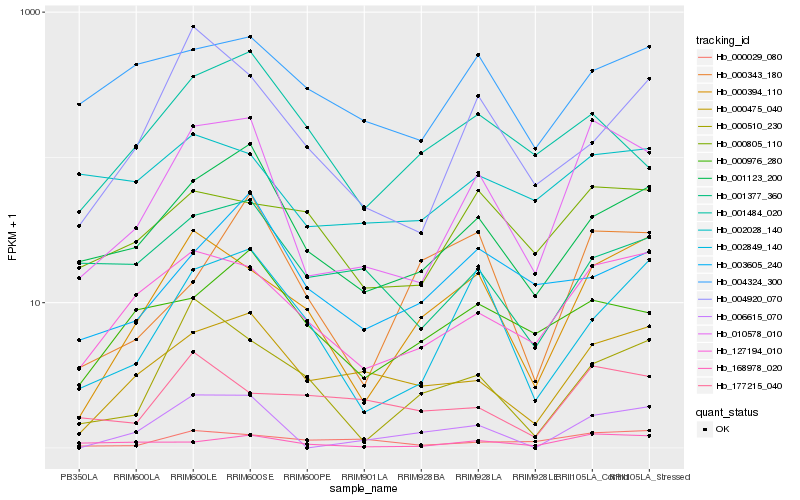
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010578_010 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 2 |
Hb_127194_010 |
0.1948096321 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 3 |
Hb_001123_200 |
0.1996502351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000475_040 |
0.2151983669 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_168978_020 |
0.2155871449 |
- |
- |
hypothetical protein POPTR_0002s10960g [Populus trichocarpa] |
| 6 |
Hb_006615_070 |
0.220113014 |
- |
- |
hypothetical protein RCOM_1466910 [Ricinus communis] |
| 7 |
Hb_000394_110 |
0.2268406874 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 8 |
Hb_002849_140 |
0.243254848 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 9 |
Hb_000029_080 |
0.248644644 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 10 |
Hb_000805_110 |
0.2486534416 |
- |
- |
PREDICTED: uncharacterized protein C24B11.05 [Jatropha curcas] |
| 11 |
Hb_000976_280 |
0.2501213722 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 12 |
Hb_000510_230 |
0.2539706309 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 17 [Jatropha curcas] |
| 13 |
Hb_001377_360 |
0.2557318845 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 14 |
Hb_002028_140 |
0.2568858776 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 15 |
Hb_004920_070 |
0.2574900775 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 16 |
Hb_004324_300 |
0.2578343252 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 17 |
Hb_001484_020 |
0.2599364138 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_003605_240 |
0.2625804197 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 19 |
Hb_000343_180 |
0.2642109958 |
- |
- |
NDWp3 [Podospora anserina] |
| 20 |
Hb_177215_040 |
0.2646486416 |
- |
- |
PREDICTED: DET1- and DDB1-associated protein 1 isoform X1 [Jatropha curcas] |