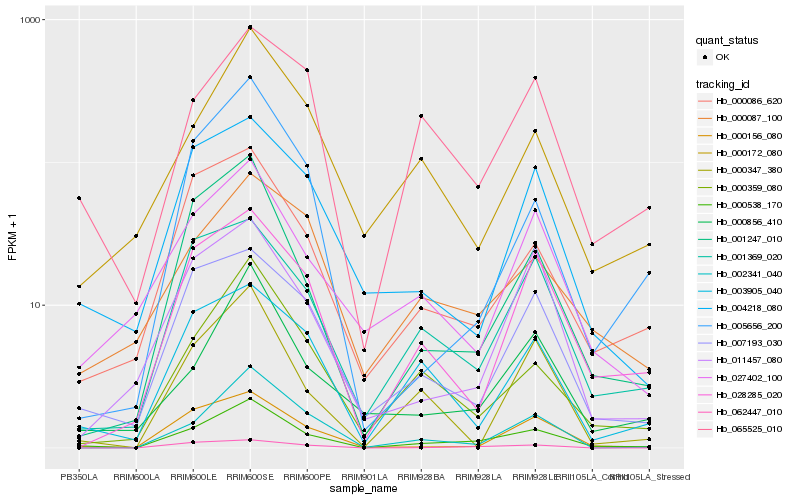
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_170 |
0.0 |
- |
- |
PREDICTED: butyrate--CoA ligase AAE11, peroxisomal-like [Jatropha curcas] |
| 2 |
Hb_001247_010 |
0.1618078156 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 3 |
Hb_000359_080 |
0.1712360933 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 4 |
Hb_001369_020 |
0.1787208567 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 5 |
Hb_000856_410 |
0.1803545597 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_002341_040 |
0.1816576109 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 7 |
Hb_000086_620 |
0.1831077412 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_028285_020 |
0.1865308812 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_007193_030 |
0.1866439018 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 10 |
Hb_065525_010 |
0.1891210738 |
- |
- |
hypothetical protein JCGZ_01481 [Jatropha curcas] |
| 11 |
Hb_062447_010 |
0.1900021768 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 12 |
Hb_000156_080 |
0.1909405289 |
- |
- |
Potassium channel KAT2, putative [Ricinus communis] |
| 13 |
Hb_003905_040 |
0.1956581121 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 14 |
Hb_000347_380 |
0.1957376267 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_027402_100 |
0.1993311524 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 16 |
Hb_000172_080 |
0.2001016628 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 17 |
Hb_000087_100 |
0.201782104 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 18 |
Hb_005656_200 |
0.201993847 |
- |
- |
Cation transport protein chaC, putative [Ricinus communis] |
| 19 |
Hb_011457_080 |
0.2039858504 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004218_080 |
0.2040916781 |
- |
- |
PREDICTED: uncharacterized protein LOC105631207 [Jatropha curcas] |