| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
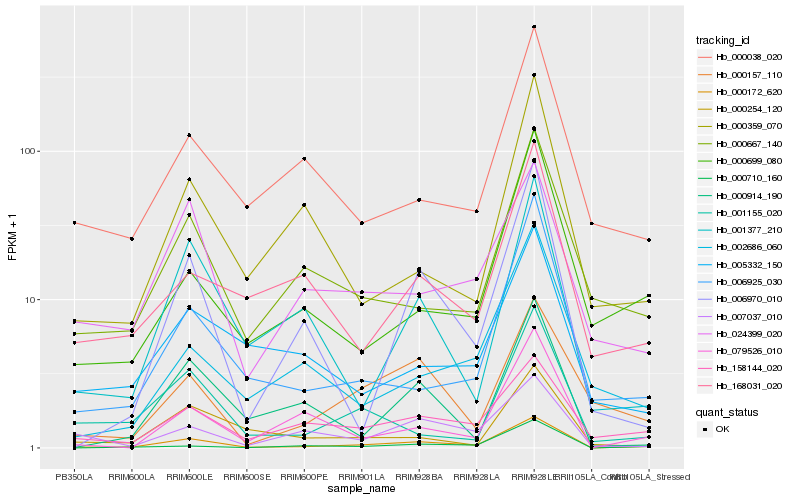
Hb_000172_620 |
0.0 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 2 |
Hb_158144_020 |
0.1896226758 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 3 |
Hb_006925_030 |
0.1967813289 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 4 |
Hb_000710_160 |
0.2036647621 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 5 |
Hb_079526_010 |
0.2043149953 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Populus euphratica] |
| 6 |
Hb_006970_010 |
0.2046533758 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 7 |
Hb_000667_140 |
0.2083286347 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001377_210 |
0.2108529475 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_000914_190 |
0.211095432 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_001155_020 |
0.214102495 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 2 [Theobroma cacao] |
| 11 |
Hb_007037_010 |
0.2145303724 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 12 |
Hb_000157_110 |
0.2170172404 |
- |
- |
hypothetical protein Csa_6G439410 [Cucumis sativus] |
| 13 |
Hb_024399_020 |
0.2177430954 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 14 |
Hb_168031_020 |
0.2183639161 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000038_020 |
0.2191936307 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000699_080 |
0.2193675359 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002686_060 |
0.2232392593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000359_070 |
0.2250229142 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005332_150 |
0.2253879752 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 20 |
Hb_000254_120 |
0.23076164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |