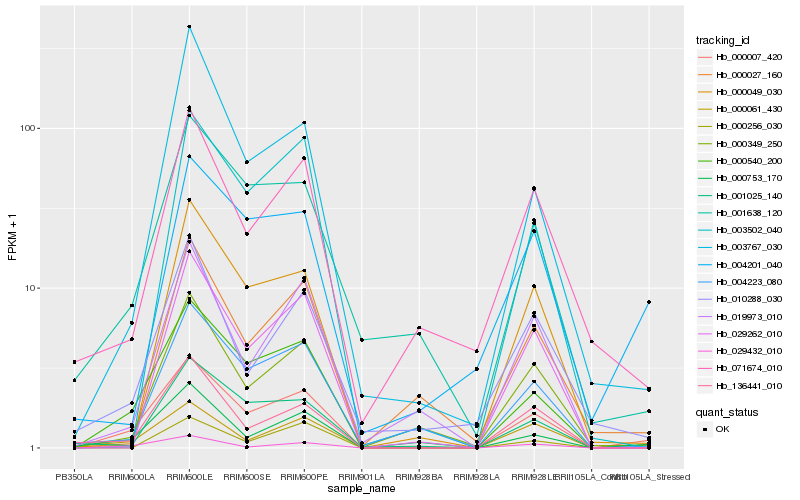
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000007_420 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 2 |
Hb_000540_200 |
0.121081687 |
- |
- |
Uncharacterized protein TCM_041994 [Theobroma cacao] |
| 3 |
Hb_000753_170 |
0.1753300429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_029432_010 |
0.1797070094 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000349_250 |
0.1801078293 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 6 |
Hb_004223_080 |
0.1849782021 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_136441_010 |
0.1862072266 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 8 |
Hb_001638_120 |
0.1869047823 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 9 |
Hb_000049_030 |
0.1939777857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000061_430 |
0.1975329126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_029262_010 |
0.1978148862 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 12 |
Hb_001025_140 |
0.2015799331 |
- |
- |
hypothetical protein POPTR_0015s09180g [Populus trichocarpa] |
| 13 |
Hb_003767_030 |
0.2040315782 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 14 |
Hb_071674_010 |
0.2043091877 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 15 |
Hb_010288_030 |
0.2044164871 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 16 |
Hb_003502_040 |
0.2052594545 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 17 |
Hb_019973_010 |
0.2079550495 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004201_040 |
0.2080460027 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 19 |
Hb_000256_030 |
0.208099498 |
- |
- |
transferase, putative [Ricinus communis] |
| 20 |
Hb_000027_160 |
0.2107984927 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |