| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
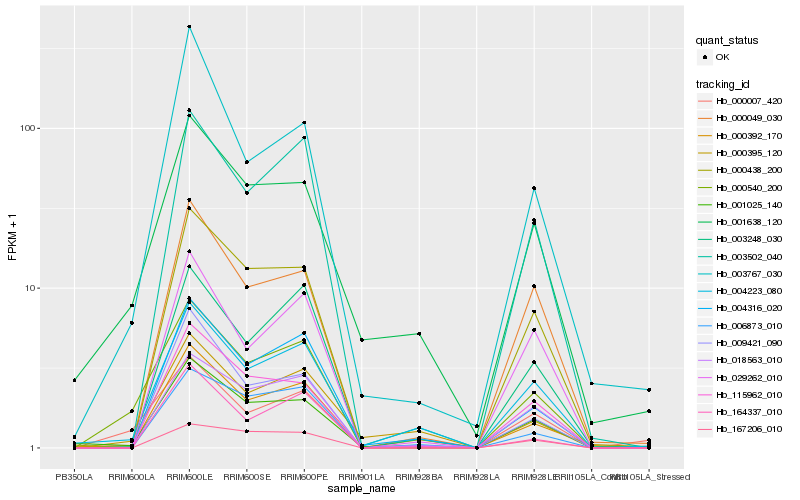
| 1 |
Hb_000540_200 |
0.0 |
- |
- |
Uncharacterized protein TCM_041994 [Theobroma cacao] |
| 2 |
Hb_000007_420 |
0.121081687 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 3 |
Hb_001025_140 |
0.1583332928 |
- |
- |
hypothetical protein POPTR_0015s09180g [Populus trichocarpa] |
| 4 |
Hb_003502_040 |
0.1647260351 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 5 |
Hb_004223_080 |
0.1649146439 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_001638_120 |
0.1661791768 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 7 |
Hb_000438_200 |
0.1666998777 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |
| 8 |
Hb_018563_010 |
0.1761768996 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 5-like [Jatropha curcas] |
| 9 |
Hb_000392_170 |
0.1767328934 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 10 |
Hb_004316_020 |
0.1771726814 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 11 |
Hb_003248_030 |
0.1784423152 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_115962_010 |
0.1802212163 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 13 |
Hb_000049_030 |
0.1831501319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000395_120 |
0.1849523569 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 15 |
Hb_029262_010 |
0.1872420578 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 16 |
Hb_009421_090 |
0.1878209764 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 17 |
Hb_003767_030 |
0.188877361 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 18 |
Hb_164337_010 |
0.1889926464 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 19 |
Hb_167206_010 |
0.1927088343 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 20 |
Hb_006873_010 |
0.1927180668 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |