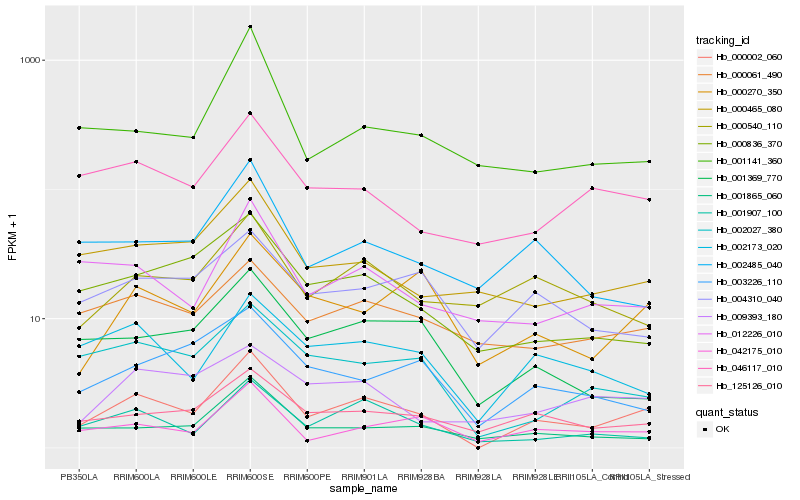
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_060 |
0.0 |
- |
- |
BnaA03g50800D [Brassica napus] |
| 2 |
Hb_125126_010 |
0.1849392173 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |
| 3 |
Hb_042175_010 |
0.1868624022 |
- |
- |
hypothetical protein CISIN_1g0016721mg, partial [Citrus sinensis] |
| 4 |
Hb_001907_100 |
0.1880078155 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 5 |
Hb_000270_350 |
0.1958246446 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 6 |
Hb_002173_020 |
0.1966815696 |
- |
- |
PREDICTED: uncharacterized protein LOC105801078 [Gossypium raimondii] |
| 7 |
Hb_001865_060 |
0.1993575768 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 8 |
Hb_002027_380 |
0.2003704035 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase isoform X2 [Jatropha curcas] |
| 9 |
Hb_012226_010 |
0.2007159903 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 10 |
Hb_046117_010 |
0.2033835633 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 11 |
Hb_001141_360 |
0.2068400896 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 12 |
Hb_000465_080 |
0.2069031608 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 13 |
Hb_001369_770 |
0.20864178 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 14 |
Hb_000836_370 |
0.2098117802 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000061_490 |
0.2098785747 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 16 |
Hb_009393_180 |
0.2115541545 |
- |
- |
PREDICTED: uncharacterized protein LOC101292333 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_003226_110 |
0.2122102328 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 18 |
Hb_000540_110 |
0.2123180786 |
- |
- |
hypothetical protein PRUPE_ppa000777mg [Prunus persica] |
| 19 |
Hb_004310_040 |
0.2126285228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002485_040 |
0.2128109375 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |