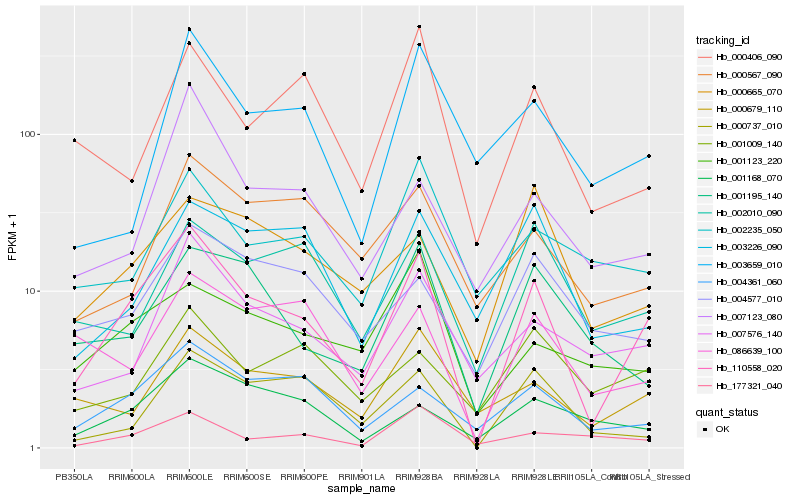
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_110558_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641876 [Jatropha curcas] |
| 2 |
Hb_004361_060 |
0.2086001064 |
- |
- |
PREDICTED: uncharacterized protein LOC105636705 [Jatropha curcas] |
| 3 |
Hb_007576_140 |
0.2108414405 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 4 |
Hb_000679_110 |
0.2175633291 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 5 |
Hb_000737_010 |
0.2305564747 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 6 |
Hb_001195_140 |
0.2340665884 |
- |
- |
PREDICTED: U-box domain-containing protein 5-like [Jatropha curcas] |
| 7 |
Hb_001168_070 |
0.2348825845 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 8 |
Hb_086639_100 |
0.235062136 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105646787 [Jatropha curcas] |
| 9 |
Hb_007123_080 |
0.2355988585 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 10 |
Hb_000665_070 |
0.2356185121 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002010_090 |
0.2362808051 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_004577_010 |
0.2374313299 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 13 |
Hb_177321_040 |
0.2386611053 |
- |
- |
hypothetical protein JCGZ_06328 [Jatropha curcas] |
| 14 |
Hb_000567_090 |
0.2396165561 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001123_220 |
0.2400052477 |
- |
- |
- |
| 16 |
Hb_001009_140 |
0.2406352511 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 17 |
Hb_000406_090 |
0.240684935 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_002235_050 |
0.2411648025 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003226_090 |
0.2412059457 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 20 |
Hb_003659_010 |
0.2417591476 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |