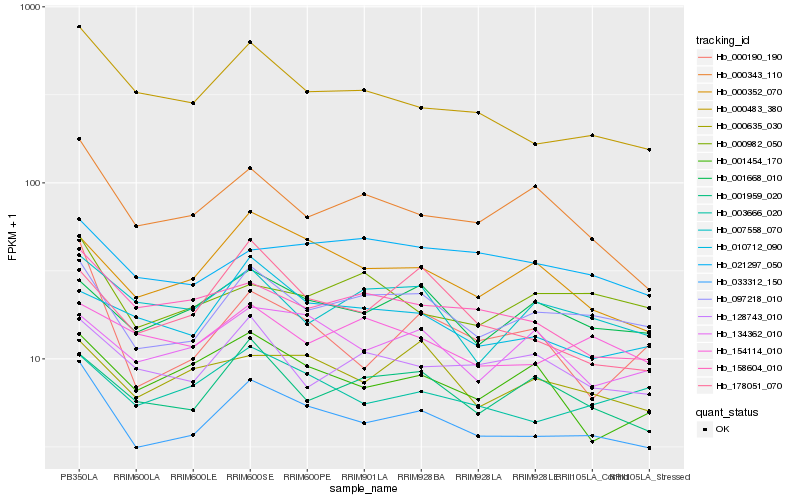
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033312_150 |
0.0 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 2 |
Hb_097218_010 |
0.080371147 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000483_380 |
0.0915260735 |
- |
- |
PREDICTED: bax inhibitor 1-like [Jatropha curcas] |
| 4 |
Hb_003666_020 |
0.1072278882 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_000352_070 |
0.1099507635 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 6 |
Hb_158604_010 |
0.1123958938 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 7 |
Hb_000343_110 |
0.1132201255 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 8 |
Hb_001668_010 |
0.1135597979 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 9 |
Hb_001959_020 |
0.1155827868 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000635_030 |
0.1169340089 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 11 |
Hb_128743_010 |
0.1194730386 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 12 |
Hb_010712_090 |
0.1198028303 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 13 |
Hb_134362_010 |
0.1223459919 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 14 |
Hb_000190_190 |
0.1235313141 |
- |
- |
PREDICTED: BI1-like protein [Gossypium raimondii] |
| 15 |
Hb_007558_070 |
0.1236801209 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 16 |
Hb_021297_050 |
0.1236973184 |
- |
- |
PREDICTED: signal recognition particle receptor subunit alpha [Jatropha curcas] |
| 17 |
Hb_000982_050 |
0.1237231493 |
- |
- |
PREDICTED: uncharacterized protein LOC105643124 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001454_170 |
0.1238984017 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 19 |
Hb_154114_010 |
0.1247223294 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 20 |
Hb_178051_070 |
0.1248251205 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |