| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033279_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein, putative [Theobroma cacao] |
| 2 |
Hb_000098_090 |
0.295291277 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 3 |
Hb_007148_010 |
0.300889153 |
- |
- |
PREDICTED: uncharacterized protein LOC103427080 [Malus domestica] |
| 4 |
Hb_012305_080 |
0.3499482441 |
- |
- |
- |
| 5 |
Hb_009986_010 |
0.3652554406 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_005714_030 |
0.3714300768 |
- |
- |
PREDICTED: COBRA-like protein 6 isoform X1 [Gossypium raimondii] |
| 7 |
Hb_008305_010 |
0.376539132 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_000463_030 |
0.3770491457 |
- |
- |
- |
| 9 |
Hb_033661_010 |
0.37758471 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 10 |
Hb_000261_300 |
0.3815335388 |
- |
- |
- |
| 11 |
Hb_001366_160 |
0.3854734251 |
- |
- |
- |
| 12 |
Hb_001021_050 |
0.3892071064 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 13 |
Hb_023226_100 |
0.3917836677 |
- |
- |
PREDICTED: uncharacterized protein LOC104903172 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_177132_010 |
0.3926519818 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_007276_020 |
0.3928427314 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_080085_010 |
0.3929968699 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 17 |
Hb_000016_120 |
0.3941115601 |
- |
- |
PREDICTED: uncharacterized protein LOC104610208 [Nelumbo nucifera] |
| 18 |
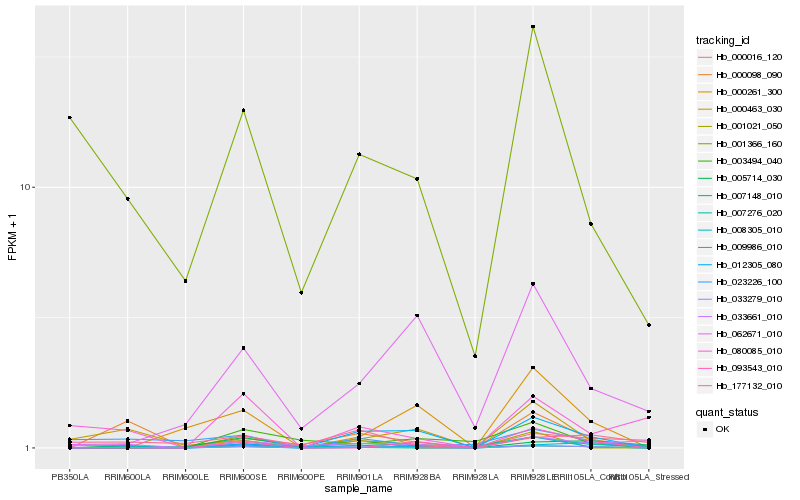
Hb_003494_040 |
0.3960755585 |
transcription factor |
TF Family: NAC |
PREDICTED: protein FEZ [Jatropha curcas] |
| 19 |
Hb_062671_010 |
0.3963568466 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_093543_010 |
0.3975359209 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |