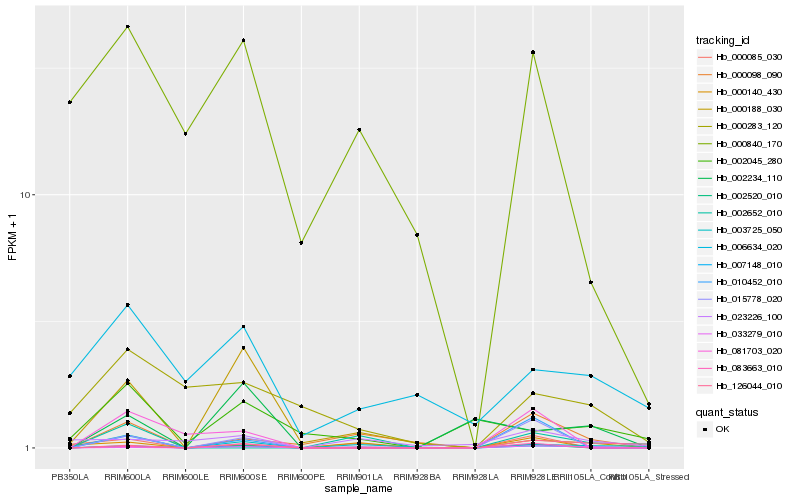
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007148_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103427080 [Malus domestica] |
| 2 |
Hb_033279_010 |
0.300889153 |
- |
- |
DNA/RNA polymerases superfamily protein, putative [Theobroma cacao] |
| 3 |
Hb_000098_090 |
0.3026796338 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 4 |
Hb_003725_050 |
0.3029596562 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_002520_010 |
0.3114047816 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 6 |
Hb_126044_010 |
0.3872059314 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 7 |
Hb_015778_020 |
0.394773535 |
- |
- |
Putative methyltransferase family protein [Solanum demissum] |
| 8 |
Hb_083663_010 |
0.398453208 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 9 |
Hb_010452_010 |
0.4163896474 |
- |
- |
PREDICTED: uncharacterized protein LOC104232369 [Nicotiana sylvestris] |
| 10 |
Hb_002234_110 |
0.4185279873 |
- |
- |
PREDICTED: uncharacterized protein LOC103455142 [Malus domestica] |
| 11 |
Hb_000085_030 |
0.4189930207 |
- |
- |
- |
| 12 |
Hb_000840_170 |
0.4297864897 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 13 |
Hb_081703_020 |
0.4305024566 |
- |
- |
- |
| 14 |
Hb_002045_280 |
0.4324460658 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_006634_020 |
0.4352726673 |
- |
- |
- |
| 16 |
Hb_023226_100 |
0.4359916786 |
- |
- |
PREDICTED: uncharacterized protein LOC104903172 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_000140_430 |
0.4370223393 |
- |
- |
- |
| 18 |
Hb_000283_120 |
0.4378456562 |
- |
- |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 19 |
Hb_002652_010 |
0.4388081777 |
- |
- |
- |
| 20 |
Hb_000188_030 |
0.4407570598 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |