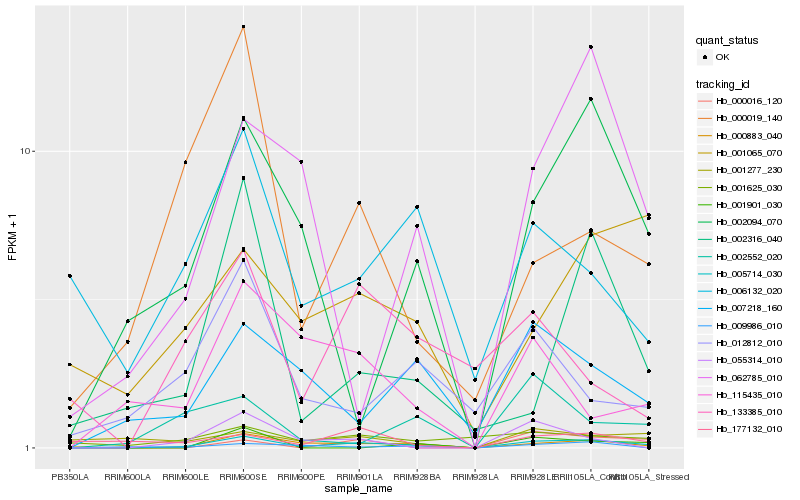
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005714_030 |
0.0 |
- |
- |
PREDICTED: COBRA-like protein 6 isoform X1 [Gossypium raimondii] |
| 2 |
Hb_000883_040 |
0.2093616373 |
- |
- |
- |
| 3 |
Hb_007218_160 |
0.3007065056 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 4 |
Hb_002094_070 |
0.3102938083 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 5 |
Hb_001065_070 |
0.3178597222 |
- |
- |
- |
| 6 |
Hb_002316_040 |
0.3254543528 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_001625_030 |
0.3261637987 |
- |
- |
hypothetical protein JCGZ_20707 [Jatropha curcas] |
| 8 |
Hb_062785_010 |
0.3303783857 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 9 |
Hb_177132_010 |
0.3317991774 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 10 |
Hb_000019_140 |
0.33187705 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 11 |
Hb_000016_120 |
0.3331600726 |
- |
- |
PREDICTED: uncharacterized protein LOC104610208 [Nelumbo nucifera] |
| 12 |
Hb_002552_020 |
0.338178276 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 13 |
Hb_006132_020 |
0.3393165437 |
- |
- |
PREDICTED: U-box domain-containing protein 37-like isoform X1 [Populus euphratica] |
| 14 |
Hb_009986_010 |
0.340237732 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_055314_010 |
0.3405915782 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_115435_010 |
0.3410485727 |
- |
- |
- |
| 17 |
Hb_012812_010 |
0.343420653 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 18 |
Hb_001901_030 |
0.3442205568 |
- |
- |
- |
| 19 |
Hb_001277_230 |
0.3450687467 |
- |
- |
- |
| 20 |
Hb_133385_010 |
0.3462422357 |
- |
- |
- |