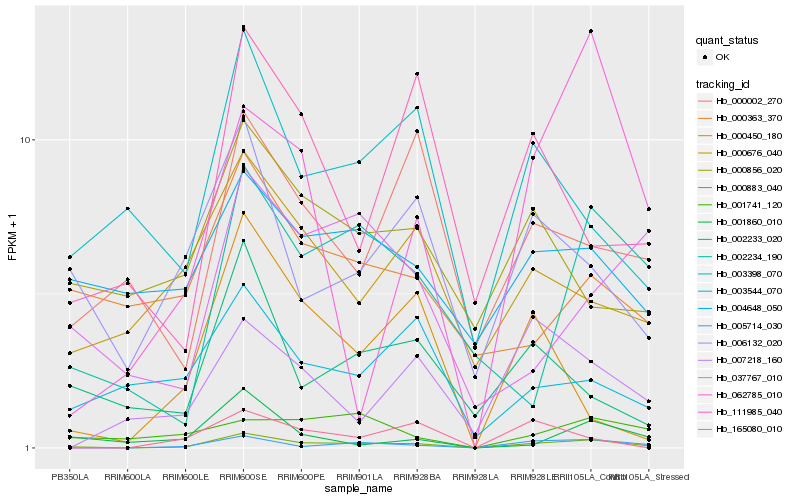
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000883_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_005714_030 |
0.2093616373 |
- |
- |
PREDICTED: COBRA-like protein 6 isoform X1 [Gossypium raimondii] |
| 3 |
Hb_002234_190 |
0.2678199715 |
- |
- |
PREDICTED: putative non-specific lipid-transfer protein 14 [Jatropha curcas] |
| 4 |
Hb_006132_020 |
0.2762026787 |
- |
- |
PREDICTED: U-box domain-containing protein 37-like isoform X1 [Populus euphratica] |
| 5 |
Hb_003398_070 |
0.2787167523 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 6 |
Hb_002233_020 |
0.2841388454 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 7 |
Hb_000002_270 |
0.2864040301 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 8 |
Hb_037767_010 |
0.2895605216 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000676_040 |
0.2906299746 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 10 |
Hb_111985_040 |
0.2922470804 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 11 |
Hb_003544_070 |
0.2931969455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001860_010 |
0.2942976819 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_007218_160 |
0.2949566516 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 14 |
Hb_001741_120 |
0.2956151951 |
- |
- |
hypothetical protein CARUB_v10011543mg, partial [Capsella rubella] |
| 15 |
Hb_004648_050 |
0.2974684521 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_000450_180 |
0.3000323349 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 17 |
Hb_165080_010 |
0.3033350085 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 18 |
Hb_000363_370 |
0.3043668222 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 19 |
Hb_062785_010 |
0.3048586871 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 20 |
Hb_000856_020 |
0.3048704352 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |