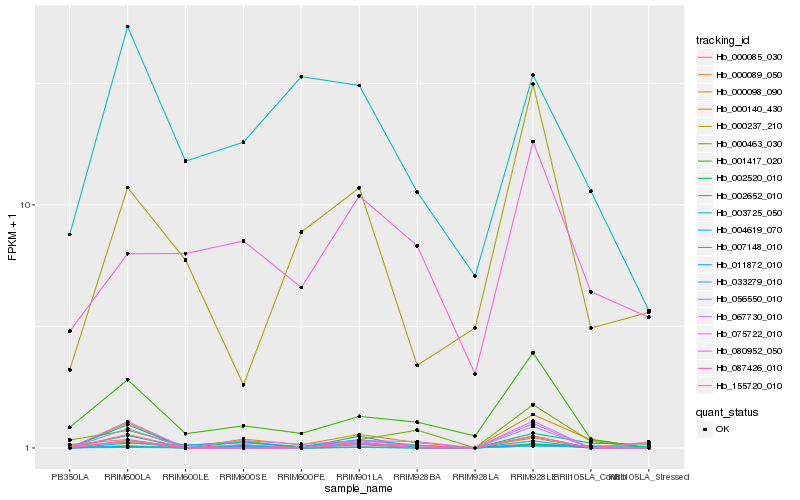
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 2 |
Hb_001417_020 |
0.2830547815 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 3 |
Hb_033279_010 |
0.295291277 |
- |
- |
DNA/RNA polymerases superfamily protein, putative [Theobroma cacao] |
| 4 |
Hb_007148_010 |
0.3026796338 |
- |
- |
PREDICTED: uncharacterized protein LOC103427080 [Malus domestica] |
| 5 |
Hb_000089_050 |
0.3056209082 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_000463_030 |
0.3171220112 |
- |
- |
- |
| 7 |
Hb_075722_010 |
0.3396462906 |
- |
- |
hypothetical protein JCGZ_01984 [Jatropha curcas] |
| 8 |
Hb_002520_010 |
0.3467278974 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 9 |
Hb_067730_010 |
0.3468219922 |
- |
- |
hypothetical protein OsJ_21419 [Oryza sativa Japonica Group] |
| 10 |
Hb_004619_070 |
0.3607415866 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 11 |
Hb_000085_030 |
0.3652557123 |
- |
- |
- |
| 12 |
Hb_087426_010 |
0.3657154327 |
- |
- |
PREDICTED: uncharacterized protein LOC105803348 [Gossypium raimondii] |
| 13 |
Hb_155720_010 |
0.3659936277 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 14 |
Hb_003725_050 |
0.3663129441 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_011872_010 |
0.3675108385 |
- |
- |
- |
| 16 |
Hb_000140_430 |
0.3682373543 |
- |
- |
- |
| 17 |
Hb_080952_050 |
0.3683344752 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Nelumbo nucifera] |
| 18 |
Hb_000237_210 |
0.3692681712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002652_010 |
0.3704205936 |
- |
- |
- |
| 20 |
Hb_056550_010 |
0.3712732572 |
- |
- |
PREDICTED: uncharacterized protein LOC104112577 [Nicotiana tomentosiformis] |