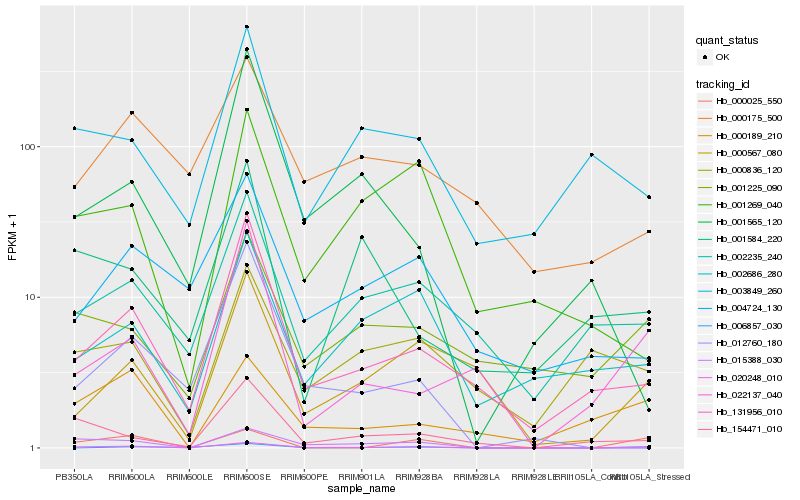
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020248_010 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_131956_010 |
0.2355410165 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 3 |
Hb_015388_030 |
0.258922828 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_154471_010 |
0.2608531863 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 5 |
Hb_022137_040 |
0.2641975911 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |
| 6 |
Hb_000189_210 |
0.2696038817 |
- |
- |
hypothetical protein CICLE_v10005534mg [Citrus clementina] |
| 7 |
Hb_000025_550 |
0.278387529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_012760_180 |
0.2831494275 |
- |
- |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 9 |
Hb_001225_090 |
0.288350264 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000567_080 |
0.2887064192 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 11 |
Hb_002235_240 |
0.2906472423 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 12 |
Hb_006857_030 |
0.2982110112 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_003849_260 |
0.298987663 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 14 |
Hb_000836_120 |
0.3020385773 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 15 |
Hb_002686_280 |
0.3023478802 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_001565_120 |
0.3029802348 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001269_040 |
0.3116161505 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000175_500 |
0.3116436757 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 19 |
Hb_001584_220 |
0.3124547452 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 20 |
Hb_004724_130 |
0.3125846793 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |