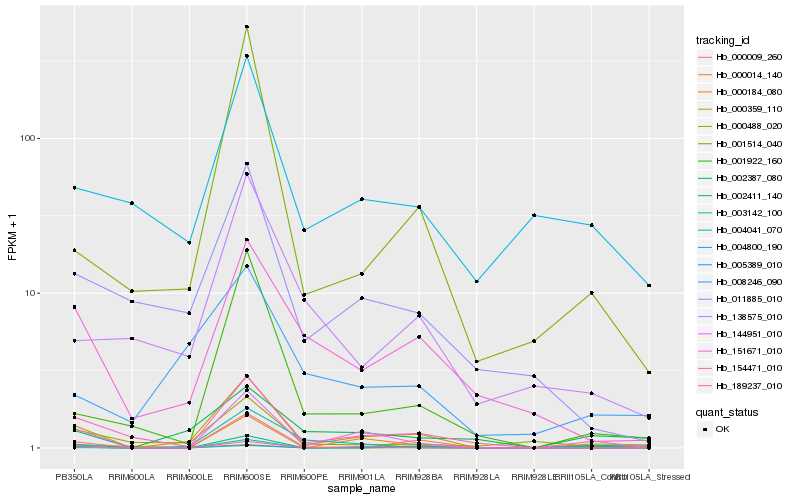
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_110 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 42 [Theobroma cacao] |
| 2 |
Hb_154471_010 |
0.2277206447 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 3 |
Hb_144951_010 |
0.2289874702 |
- |
- |
putative reverse transcriptase [Oryza sativa Japonica Group] |
| 4 |
Hb_005389_010 |
0.2481119392 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 5 |
Hb_001514_040 |
0.2505122886 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 6 |
Hb_001922_160 |
0.2626302257 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 7 |
Hb_000184_080 |
0.2640508257 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 8 |
Hb_011885_010 |
0.2649996471 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 9 |
Hb_189237_010 |
0.2653041152 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 10 |
Hb_000009_260 |
0.2741825496 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002411_140 |
0.2745190303 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Jatropha curcas] |
| 12 |
Hb_000488_020 |
0.2747348831 |
- |
- |
PREDICTED: UDP-glucose iridoid glucosyltransferase-like [Jatropha curcas] |
| 13 |
Hb_003142_100 |
0.2753469508 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Eucalyptus grandis] |
| 14 |
Hb_000014_140 |
0.2782659965 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 15 |
Hb_151671_010 |
0.2786099245 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 16 |
Hb_004041_070 |
0.2788394772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004800_190 |
0.2825920938 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 18 |
Hb_002387_080 |
0.2838285611 |
- |
- |
hypothetical protein JCGZ_25275 [Jatropha curcas] |
| 19 |
Hb_008246_090 |
0.2840777048 |
- |
- |
- |
| 20 |
Hb_138575_010 |
0.2849516577 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |