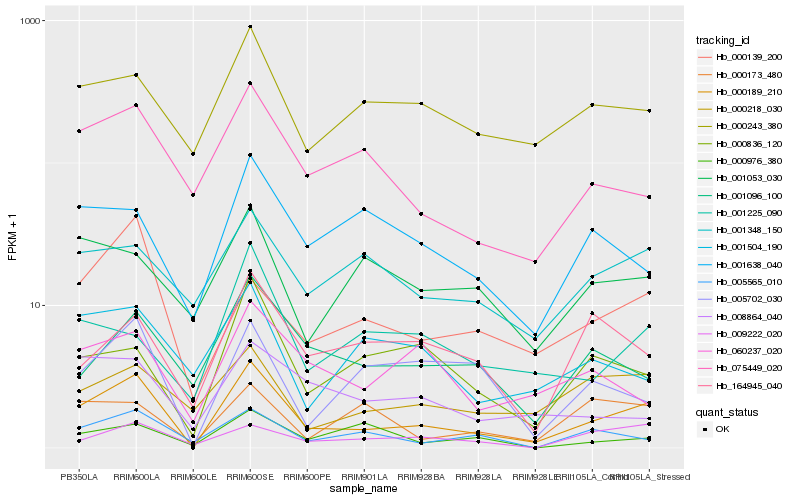
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_210 |
0.0 |
- |
- |
hypothetical protein CICLE_v10005534mg [Citrus clementina] |
| 2 |
Hb_075449_020 |
0.2285204739 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 3 |
Hb_000836_120 |
0.2324065555 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_000218_030 |
0.236643869 |
- |
- |
- |
| 5 |
Hb_000139_200 |
0.2370059303 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_005565_010 |
0.2375561644 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 7 |
Hb_000243_380 |
0.2382925361 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 8 |
Hb_001096_100 |
0.2428980819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009222_020 |
0.2433281793 |
- |
- |
hypothetical protein AALP_AA5G087000 [Arabis alpina] |
| 10 |
Hb_001348_150 |
0.2480015468 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005702_030 |
0.2483045911 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 12 |
Hb_164945_040 |
0.2487763982 |
- |
- |
PREDICTED: uncharacterized protein LOC105628261 [Jatropha curcas] |
| 13 |
Hb_001638_040 |
0.2508558396 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 14 |
Hb_060237_020 |
0.2522974966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001225_090 |
0.2542396584 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_008864_040 |
0.2564995421 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 17 |
Hb_001504_190 |
0.2593619706 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_001053_030 |
0.2611572925 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000173_480 |
0.2621856777 |
- |
- |
PREDICTED: calcium-binding protein 39 [Jatropha curcas] |
| 20 |
Hb_000976_380 |
0.2628804137 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |