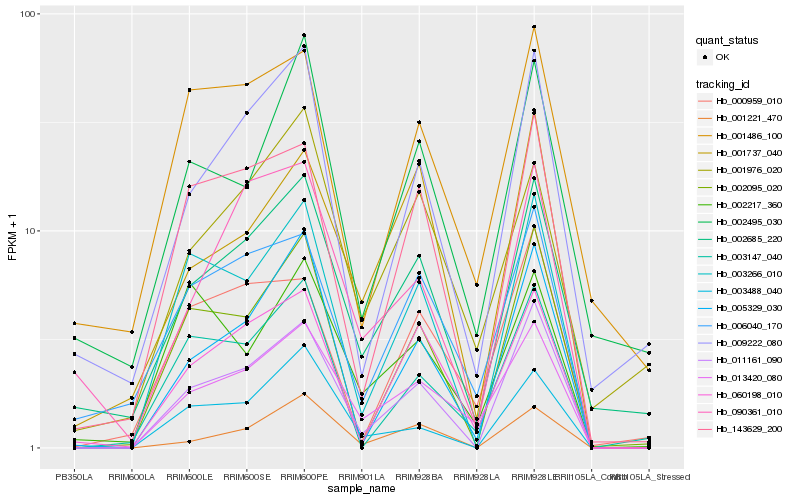
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013420_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011161_090 |
0.1481078537 |
- |
- |
- |
| 3 |
Hb_002685_220 |
0.1559307545 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_006040_170 |
0.1565331969 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 5 |
Hb_060198_010 |
0.1583442112 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_001976_020 |
0.1617214669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003266_010 |
0.1744784449 |
- |
- |
hypothetical protein JCGZ_21945 [Jatropha curcas] |
| 8 |
Hb_005329_030 |
0.1752064158 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 9 |
Hb_009222_080 |
0.1803206558 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 10 |
Hb_001221_470 |
0.181884502 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 11 |
Hb_003488_040 |
0.1819904004 |
transcription factor |
TF Family: NF-YB |
nuclear transcription factor YB7 [Populus nigra x Populus x canadensis] |
| 12 |
Hb_090361_010 |
0.1822976574 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_001737_040 |
0.1824683942 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 14 |
Hb_143629_200 |
0.1831682126 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 15 |
Hb_002095_020 |
0.1859683833 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000959_010 |
0.1876972393 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003147_040 |
0.1896727895 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 18 |
Hb_001486_100 |
0.1903105242 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 19 |
Hb_002217_360 |
0.1915031079 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002495_030 |
0.1925226554 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |