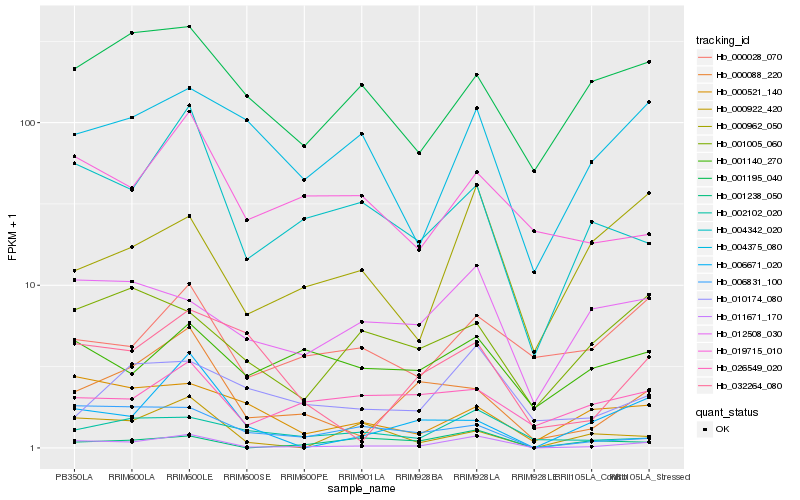
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_170 |
0.0 |
- |
- |
PREDICTED: protein DYAD [Jatropha curcas] |
| 2 |
Hb_004342_020 |
0.2409589554 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000088_220 |
0.2487983246 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_012508_030 |
0.2685323646 |
- |
- |
PREDICTED: uncharacterized protein LOC105636284 [Jatropha curcas] |
| 5 |
Hb_000962_050 |
0.2692132954 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 6 |
Hb_002102_020 |
0.2724387505 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 7 |
Hb_001238_050 |
0.2756006816 |
- |
- |
- |
| 8 |
Hb_006831_100 |
0.2759340393 |
- |
- |
snRNA-activating protein complex subunit, putative [Ricinus communis] |
| 9 |
Hb_019715_010 |
0.2818682833 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 10 |
Hb_000922_420 |
0.2821852847 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 11 |
Hb_004375_080 |
0.2829515566 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 12 |
Hb_006671_020 |
0.2832239304 |
- |
- |
hypothetical protein CISIN_1g0407181mg, partial [Citrus sinensis] |
| 13 |
Hb_032264_080 |
0.2850825729 |
- |
- |
PREDICTED: uncharacterized protein LOC105648216 [Jatropha curcas] |
| 14 |
Hb_000028_070 |
0.285795453 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 15 |
Hb_001195_040 |
0.2864777172 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 16 |
Hb_010174_080 |
0.2874420496 |
- |
- |
- |
| 17 |
Hb_000521_140 |
0.2889561115 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 18 |
Hb_026549_020 |
0.291384436 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_001005_060 |
0.2944667999 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase MAK3 [Jatropha curcas] |
| 20 |
Hb_001140_270 |
0.2956847611 |
- |
- |
hypothetical protein EUGRSUZ_E00166 [Eucalyptus grandis] |