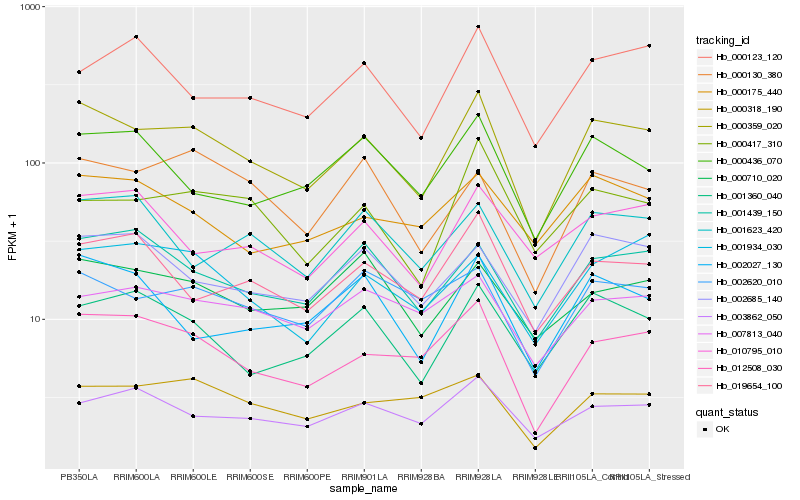
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_020 |
0.0 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 2 |
Hb_012508_030 |
0.0940197597 |
- |
- |
PREDICTED: uncharacterized protein LOC105636284 [Jatropha curcas] |
| 3 |
Hb_001439_150 |
0.111342858 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001360_040 |
0.1123401448 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002620_010 |
0.1147142097 |
- |
- |
hypothetical protein POPTR_0004s18130g [Populus trichocarpa] |
| 6 |
Hb_002685_140 |
0.1158684444 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 7 |
Hb_019654_100 |
0.11964496 |
- |
- |
PREDICTED: tRNA pseudouridine synthase A, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_003862_050 |
0.1206306466 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105641042 [Jatropha curcas] |
| 9 |
Hb_000417_310 |
0.121773593 |
- |
- |
uncharacterized protein LOC100272531 [Zea mays] |
| 10 |
Hb_000175_440 |
0.121794509 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 11 |
Hb_002027_130 |
0.121925114 |
- |
- |
PREDICTED: uncharacterized protein LOC105650052 [Jatropha curcas] |
| 12 |
Hb_000130_380 |
0.1220521083 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001934_030 |
0.1238676656 |
transcription factor |
TF Family: HSF |
Heat shock factor protein, putative [Ricinus communis] |
| 14 |
Hb_000318_190 |
0.1240827487 |
- |
- |
hypothetical protein JCGZ_04889 [Jatropha curcas] |
| 15 |
Hb_000710_020 |
0.1241603705 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MARCH2 isoform X2 [Populus euphratica] |
| 16 |
Hb_000436_070 |
0.1243184286 |
- |
- |
PREDICTED: V-type proton ATPase subunit B 2 [Jatropha curcas] |
| 17 |
Hb_007813_040 |
0.1257428295 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 34 isoform X2 [Jatropha curcas] |
| 18 |
Hb_010795_010 |
0.1278332604 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_001623_420 |
0.1284025418 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 20 |
Hb_000123_120 |
0.1288718941 |
- |
- |
actin depolymerizing factor 4 [Hevea brasiliensis] |